2BZG
 
 | | Crystal structure of thiopurine S-methyltransferase. | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, THIOPURINE S-METHYLTRANSFERASE | | Authors: | Battaile, K.P, Wu, H, Zeng, H, Loppnau, P, Dong, A, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-25 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Allele Variation of Human Thiopurine-S-Methyltransferase.
Proteins, 67, 2007
|
|
1IF4
 
 | |
2C4F
 
 | | crystal structure of factor VII.stf complexed with pd0297121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[6-{3-[AMINO(IMINO)METHYL]PHENOXY}-4-(DIISOPROPYLAMINO)-3,5-DIFLUOROPYRIDIN-2-YL]OXY}-5-[(ISOBUTYLAMINO)CARBONYL]BEN ZOIC ACID, CALCIUM ION, ... | | Authors: | Kohrt, J.T, Zhang, E. | | Deposit date: | 2005-10-18 | | Release date: | 2006-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The Discovery of Fluoropyridine-Based Inhibitors of the Factor Viia/Tf Complex--Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ISM
 
 | | Crystal Structure Analysis of BST-1/CD157 complexed with nicotinamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NICOTINAMIDE, bone marrow stromal cell antigen 1 | | Authors: | Yamamoto-Katayama, S, Ariyoshi, M, Ishihara, K, Hirano, T, Jingami, H, Morikawa, K. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities.
J.Mol.Biol., 316, 2002
|
|
1MWE
 
 | |
2Q2A
 
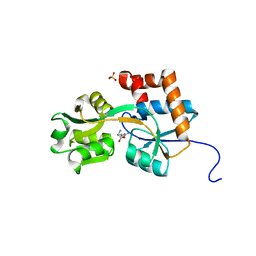 | | Crystal structures of the arginine-, lysine-, histidine-binding protein ArtJ from the thermophilic bacterium Geobacillus stearothermophilus | | Descriptor: | ARGININE, ArtJ, SULFATE ION | | Authors: | Vahedi-Faridi, A, Scheffel, F, Eckey, V, Saenger, W, Schneider, E. | | Deposit date: | 2007-05-26 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2
J.Mol.Biol., 375, 2008
|
|
2BFU
 
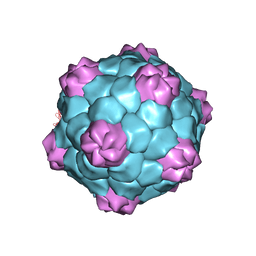 | | X-ray structure of CPMV top component | | Descriptor: | COWPEA MOSAIC VIRUS, LARGE (L) SUBUNIT, SMALL (S) SUBUNIT | | Authors: | Ochoa, W.F, Chatterji, A, Lin, T, Johnson, J.E. | | Deposit date: | 2004-12-13 | | Release date: | 2005-01-12 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Generation and Structural Analysis of Reactive Empty Particles Derived from an Icosahedral Virus.
Chem.Biol., 13, 2006
|
|
1NJE
 
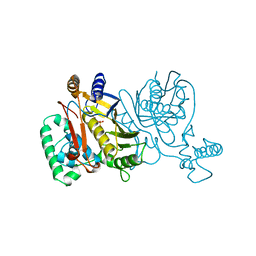 | |
1J3Z
 
 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Fe-CO)-beta(Ni) hemoglobin (laser unphotolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1ITF
 
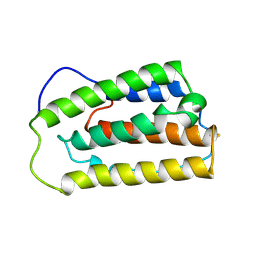 | | INTERFERON ALPHA-2A, NMR, 24 STRUCTURES | | Descriptor: | INTERFERON ALPHA-2A | | Authors: | Klaus, W, Gsell, B, Labhardt, A.M, Wipf, B, Senn, H. | | Deposit date: | 1997-08-22 | | Release date: | 1997-12-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional high resolution structure of human interferon alpha-2a determined by heteronuclear NMR spectroscopy in solution.
J.Mol.Biol., 274, 1997
|
|
2BI1
 
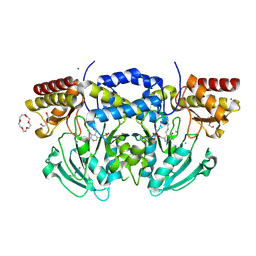 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure B) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI5
 
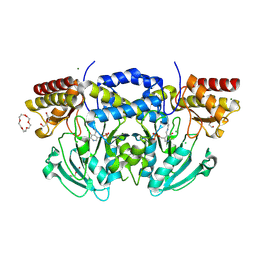 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure E) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
1J8E
 
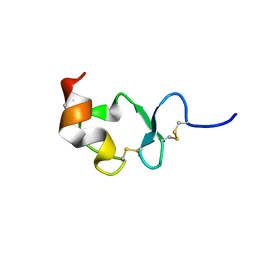 | | Crystal structure of ligand-binding repeat CR7 from LRP | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR-RELATED PROTEIN 1 | | Authors: | Simonovic, M, Dolmer, K, Huang, W, Strickland, D.K, Volz, K, Gettins, P.G.W. | | Deposit date: | 2001-05-21 | | Release date: | 2001-12-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor.
Biochemistry, 40, 2001
|
|
1NC9
 
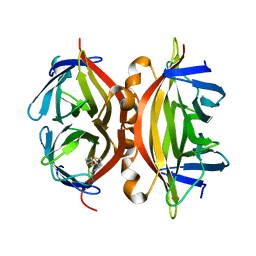 | | STREPTAVIDIN MUTANT Y43A WITH IMINOBIOTIN AT 1.8A RESOLUTION | | Descriptor: | 2-IMINOBIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-05 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2C1Z
 
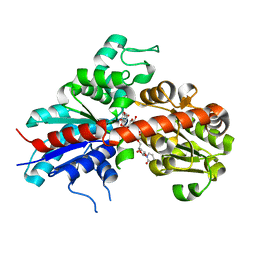 | | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, UDP-GLUCOSE FLAVONOID 3-O GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-09-22 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
1NJP
 
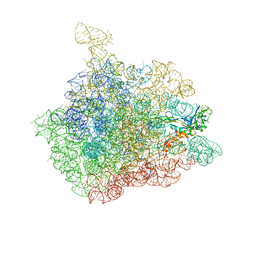 | | The crystal structure of the 50S Large ribosomal subunit from Deinococcus radiodurans complexed with a tRNA acceptor stem mimic (ASM) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L16, GENERAL STRESS PROTEIN CTC, ... | | Authors: | Bashan, A, Agmon, I, Zarivatch, R, Schluenzen, F, Harms, J.M, Berisio, R, Bartels, H, Hansen, H.A, Yonath, A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of the ribosomal machinery for Peptide bond formation,
translocation, and nascent chain progression
Mol.Cell, 11, 2003
|
|
1NI2
 
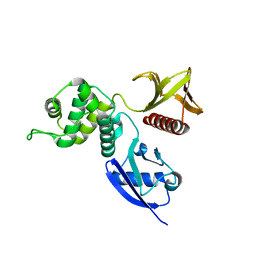 | | Structure of the active FERM domain of Ezrin | | Descriptor: | Ezrin | | Authors: | Smith, W.J, Nassar, N, Bretscher, A.P, Cerione, R.A, Karplus, P.A. | | Deposit date: | 2002-12-20 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Active N-terminal Domain of Ezrin. CONFORMATIONAL AND MOBILITY CHANGES IDENTIFY KEYSTONE INTERACTIONS.
J.Biol.Chem., 278, 2003
|
|
1NIV
 
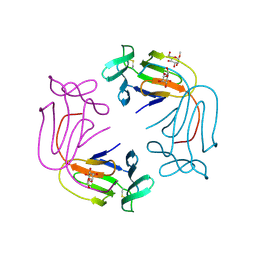 | |
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2BVL
 
 | | Crystal structure of the catalytic domain of toxin B from Clostridium difficile in complex with UDP, Glc and manganese ion | | Descriptor: | HEXATANTALUM DODECABROMIDE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Reinert, D.J, Jank, T, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Function of Clostridium Difficile Toxin B.
J.Mol.Biol., 351, 2005
|
|
2BVV
 
 | |
2BYL
 
 | | Structure of ornithine aminotransferase triple mutant Y85I Y55A G320F | | Descriptor: | ORNITHINE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Markova, M, Peneff, C, Hewlins, M.J.E, Schirmer, T, John, R.A. | | Deposit date: | 2005-08-03 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Determinants of Substrate Specificity in Omega-Aminotransferases.
J.Biol.Chem., 280, 2005
|
|
2C36
 
 | | Structure of unliganded HSV gD reveals a mechanism for receptor- mediated activation of virus entry | | Descriptor: | CHLORIDE ION, GLYCOPROTEIN D HSV-1, ZINC ION, ... | | Authors: | Krummenacher, C, Supekar, V.M, Whitbeck, J.C, Lazear, E, Connolly, S.A, Eisenberg, R.J, Cohen, G.H, Wiley, D.C, Carfi, A. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry.
EMBO J., 24, 2005
|
|
1NL1
 
 | | BOVINE PROTHROMBIN FRAGMENT 1 IN COMPLEX WITH CALCIUM ION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, M, Huang, G, Furie, B, Seaton, B, Furie, B.C. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins.
Nat.Struct.Biol., 10, 2003
|
|
1NDJ
 
 | | Streptavidin Mutant Y43F with Biotin at 1.81A Resolution | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-09 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
