6QS6
 
 | |
9G0B
 
 | |
8XOJ
 
 | | Cryo-EM structure of GPR30-Gq complex structure in the presence of G-1 | | Descriptor: | G-protein coupled estrogen receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Xu, P, Xu, H.E. | | Deposit date: | 2024-01-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional evidence that GPR30 is not a direct estrogen receptor.
Cell Res., 34, 2024
|
|
9RNT
 
 | |
6QM5
 
 | | Cryo-EM structure of calcium-bound nhTMEM16 lipid scramblase in DDM | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Kalienkova, V, Clerico Mosina, V, Bryner, L, Oostergetel, G.T, Dutzler, R, Paulino, C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Elife, 8, 2019
|
|
8WXF
 
 | | PNPase of Mycobacterium tuberculosis | | Descriptor: | Bifunctional guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | | Authors: | Wang, N, Sheng, Y.N, Liu, Y.T. | | Deposit date: | 2023-10-29 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis polynucleotide phosphorylase suggest a potential mechanism for its RNA substrate degradation.
Arch.Biochem.Biophys., 754, 2024
|
|
4TV9
 
 | | Tubulin-PM060184 complex | | Descriptor: | (1Z,4S,6Z)-1-[(N-{(2Z,4Z,6E,8S)-8-[(2S)-5-methoxy-6-oxo-3,6-dihydro-2H-pyran-2-yl]-6-methylnona-2,4,6-trienoyl}-3-methy l-L-valyl)amino]octa-1,6-dien-4-yl carbamate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Bargsten, K, Diaz, J.F, Marsh, M, Cuevas, C, Liniger, M, Neuhaus, C, Andreu, J.M, Altmann, K.H, Steinmetz, M.O. | | Deposit date: | 2014-06-26 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new tubulin-binding site and pharmacophore for microtubule-destabilizing anticancer drugs.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
9ASB
 
 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
8VL3
 
 | |
4TW6
 
 | | The Fk1 domain of FKBP51 in complex with iFit1 | | Descriptor: | (3-{(1R)-3-(3,4-dimethoxyphenyl)-1-[({(2S)-1-[(2S)-2-(3,4,5-trimethoxyphenyl)pent-4-enoyl]piperidin-2-yl}carbonyl)oxy]propyl}phenoxy)acetic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8YBK
 
 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 2024
|
|
4TW8
 
 | | The Fk1-Fk2 domains of FKBP52 in complex with iFit-FL | | Descriptor: | 2-(5-{[({3-[(1R)-1-[({(2S)-1-[(2S)-2-[(1S)-cyclohex-2-en-1-yl]-2-(3,4,5-trimethoxyphenyl)acetyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetyl)amino]methyl}-6-hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid, Peptidyl-prolyl cis-trans isomerase FKBP4 | | Authors: | Gaali, S, Kirschner, A, Cuboni, S, Hartmann, J, Kozany, C, Balsevich, G, Namendorf, C, Fernandez-Vizarra, P, Almeida, O.F.X, Ruehter, G, Uhr, M, Schmidt, M.V, Touma, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-06-30 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Selective inhibitors of the FK506-binding protein 51 by induced fit.
Nat.Chem.Biol., 11, 2015
|
|
8XQT
 
 | | Structure of human class T GPCR TAS2R14-Gi complex. | | Descriptor: | CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hu, X.L, Pei, Y, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
8WLM
 
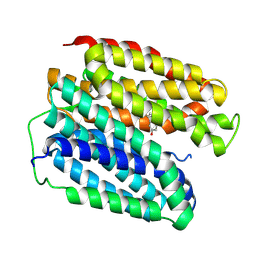 | |
9C7V
 
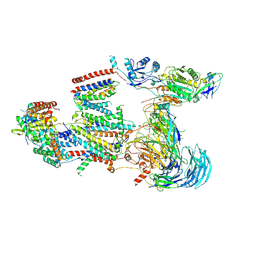 | | Structure of the human BOS:human EMC complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 2024
|
|
8YV8
 
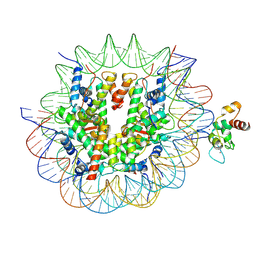 | | Cryo-EM structure of CDCA7 bound to nucleosome including hemimethylated CpG site in Widom601 positioning sequence. | | Descriptor: | Cell division cycle-associated protein 7, DNA (132-MER), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, A, Shikimachi, R, Nishiyama, A, Funabiki, H, Arita, K. | | Deposit date: | 2024-03-28 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CDCA7 is a hemimethylated DNA adaptor for the nucleosome remodeler HELLS
To Be Published
|
|
7S0T
 
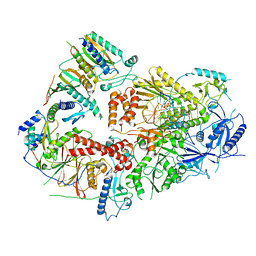 | | Structure of DNA polymerase zeta with mismatched DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Malik, R, Ubarretxena, I.B, Aggarwal, A.K. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structure of translesion DNA synthesis polymerase zeta with a base pair mismatch.
Nat Commun, 13, 2022
|
|
7RYC
 
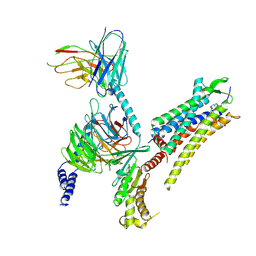 | | Oxytocin receptor (OTR) bound to oxytocin in complex with a heterotrimeric Gq protein | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(i) subunit alpha-2,Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Meyerowitz, J.G, Robertson, M.J, Skiniotis, G. | | Deposit date: | 2021-08-24 | | Release date: | 2022-03-09 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The oxytocin signaling complex reveals a molecular switch for cation dependence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8XIU
 
 | | Cryo-EM structure of a frog VMAT2 in an apo conformation | | Descriptor: | XlVMAT2 | | Authors: | Lyu, Y, Fu, C, Ma, H, Sun, Z, Su, Z, Zhou, X. | | Deposit date: | 2023-12-20 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Engineering of a mammalian VMAT2 for cryo-EM analysis results in non-canonical protein folding.
Nat Commun, 15, 2024
|
|
4TYE
 
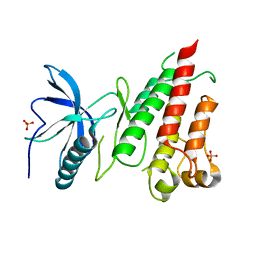 | | Structural analysis of the human Fibroblast Growth Factor Receptor 4 Kinase | | Descriptor: | Fibroblast growth factor receptor 4, PHOSPHATE ION | | Authors: | Lesca, E, Lammens, A, Huber, R, Augustin, M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the human fibroblast growth factor receptor 4 kinase.
J.Mol.Biol., 426, 2014
|
|
8XQO
 
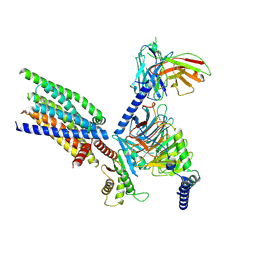 | | Structure of human class T GPCR TAS2R14-Gi complex with Aristolochic acid A. | | Descriptor: | 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, CHOLESTEROL, Exo-alpha-sialidase,Taste receptor type 2 member 14,LgBiT, ... | | Authors: | Hu, X.L, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Bitter taste TAS2R14 activation by intracellular tastants and cholesterol.
Nature, 631, 2024
|
|
9B3D
 
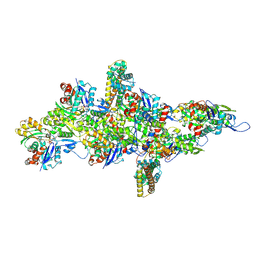 | | mDia1 in the middle of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Palmer, N.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2024-03-19 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanisms of actin filament severing and elongation by formins.
Nature, 632, 2024
|
|
8YKV
 
 | | Cryo-EM structure of succinate receptor SUCR1 bound to compound 31 | | Descriptor: | (2~{S})-2-[[6-[4-(trifluoromethyloxy)phenyl]pyridin-2-yl]carbonylamino]butanedioic acid, Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Li, C, Liu, H, Li, J, Zhu, H, Fu, W, Xu, H.E. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Molecular basis of ligand recognition and activation of the human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
9COJ
 
 | | SH3-like tandem domain of human KIN protein | | Descriptor: | DNA/RNA-binding protein KIN17 | | Authors: | de Lourenco, I.O, Fossey, M.A, Souza, F.P, Seixas, F.A.V, Fernandez, M.A, Almeida, F.C.L, Caruso, I.P. | | Deposit date: | 2024-07-16 | | Release date: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SH3-like tandem domain of human KIN protein and its interaction with RNA homo-oligonucleotides
To Be Published
|
|
