5XM2
 
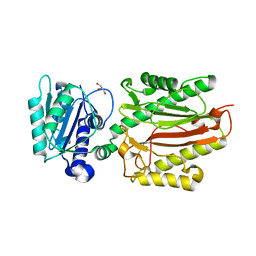 | | Human N-terminal domain of FACT complex subunit SPT16 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FACT complex subunit SPT16, GLYCEROL | | Authors: | Xu, S, Li, H, Dou, Y, Chen, Y, Jiang, H, Lu, D, Wang, M, Su, D. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structural basis of human Spt16 N-terminal domain interaction with histone (H3-H4)2tetramer.
Biochem.Biophys.Res.Commun., 508, 2019
|
|
5XYB
 
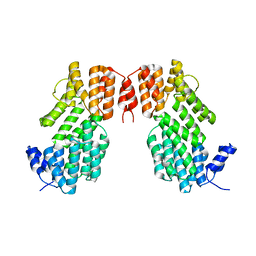 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | Descriptor: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | Authors: | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | Deposit date: | 2017-07-24 | | Release date: | 2018-09-19 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
5A7R
 
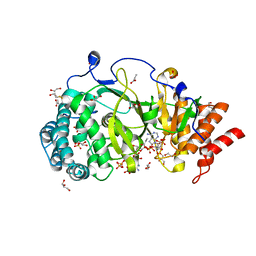 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | Deposit date: | 2015-07-09 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
5AGQ
 
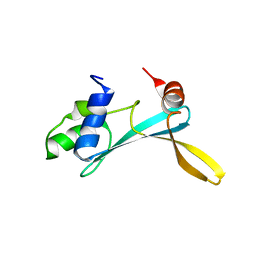 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | Descriptor: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | Authors: | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | Deposit date: | 2015-02-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
5YB1
 
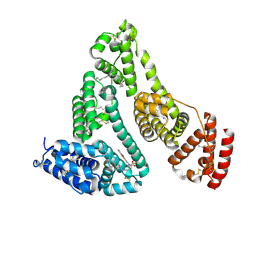 | | Structure and function of human serum albumin-metal agent complex | | Descriptor: | PALMITIC ACID, Serum albumin, ~{N},~{N},12-trimethyl-3$l^{3}-thia-1$l^{4},5,6$l^{4}-triaza-2$l^{3}-cupratricyclo[6.4.0.0^{2,6}]dodeca-1(8),3,6,9,11-pentaen-4-amine | | Authors: | Yang, F, Wang, T, Wang, J, Gou, Y, Zhang, Z.L. | | Deposit date: | 2017-09-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Developing an Anticancer Copper(II) Multitarget Pro-Drug Based on the His146 Residue in the IB Subdomain of Modified Human Serum Albumin.
Mol. Pharm., 15, 2018
|
|
4ZXQ
 
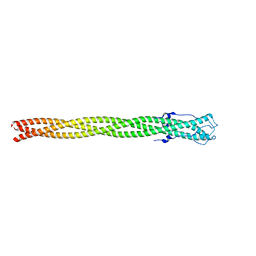 | | P22 Tail Needle Gp26 1-140 crystallized at pH 3.9 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tail needle protein gp26 | | Authors: | Sankhala, R.S, Cingolani, G. | | Deposit date: | 2015-05-20 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Plasticity of the Protein Plug That Traps Newly Packaged Genomes in Podoviridae Virions.
J.Biol.Chem., 291, 2016
|
|
280D
 
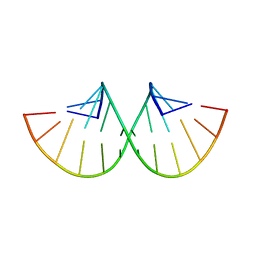 | |
5BON
 
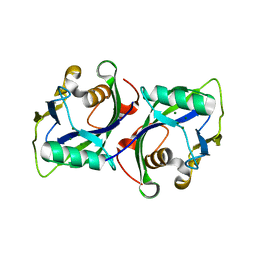 | | Crystal structure of human Nudt15 (MTH2) | | Descriptor: | MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | Authors: | Carter, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2.
Nat Commun, 6, 2015
|
|
7YPI
 
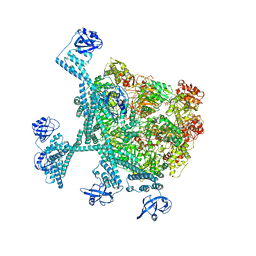 | | Spiral hexamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUH
 
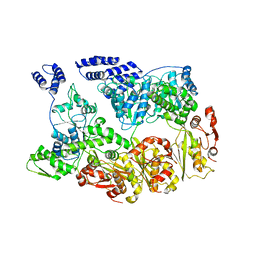 | | MtaLon-Apo for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPJ
 
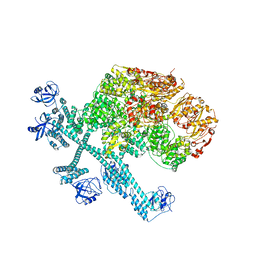 | | Spiral pentamer of the substrate-free Lon protease with a S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUU
 
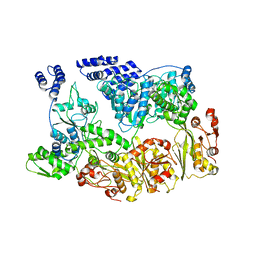 | | MtaLon-ADP for the spiral oligomers of trimer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUP
 
 | | MtaLon-Apo for the spiral oligomers of pentamer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPH
 
 | | Open-spiral pentamer of the substrate-free Lon protease with a Y224S mutation | | Descriptor: | Lon protease, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUT
 
 | | MtaLon-Apo for the spiral oligomers of hexamer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUV
 
 | | MtaLon-ADP for the spiral oligomers of tetramer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUW
 
 | | MtaLon-ADP for the spiral oligomers of pentamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YPK
 
 | | Close-ring hexamer of the substrate-bound Lon protease with an S678A mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease, alpha-S1-casein | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Lee, S.H, Ho, M.R, Wang, C.H, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUM
 
 | | MtaLon-Apo for the spiral oligomers of tetramer | | Descriptor: | Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-17 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
7YUX
 
 | | MtaLon-ADP for the spiral oligomers of hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Ho, M, Wang, C, Zhang, K, Chang, C.I. | | Deposit date: | 2022-08-18 | | Release date: | 2023-10-25 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
1JI4
 
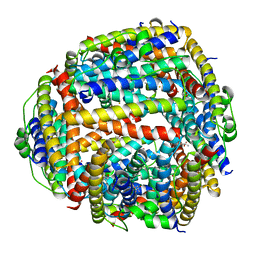 | | NAP protein from helicobacter pylori | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, NEUTROPHIL-ACTIVATING PROTEIN A, ... | | Authors: | Zanotti, G, Papinutto, E, Dundon, W.G, Battistutta, R, Seveso, M, Del Giudice, G, Rappuoli, R, Montecucco, C. | | Deposit date: | 2001-06-29 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Neutrophil-activating Protein from Helicobacter pylori
J.Mol.Biol., 323, 2002
|
|
1U0J
 
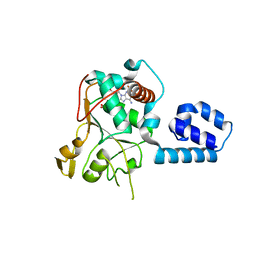 | | Crystal Structure of AAV2 Rep40-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication protein | | Authors: | James, J.A, Aggarwal, A.K, Linden, R.M, Escalante, C.R. | | Deposit date: | 2004-07-13 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of adeno-associated virus type 2 Rep40-ADP complex: Insight into
nucleotide recognition and catalysis by superfamily 3 helicases
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TPT
 
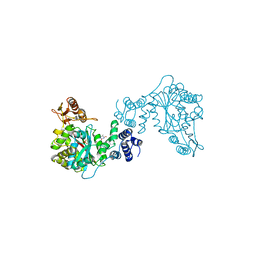 | | THREE-DIMENSIONAL STRUCTURE OF THYMIDINE PHOSPHORYLASE FROM ESCHERICHIA COLI AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | SULFATE ION, THYMIDINE PHOSPHORYLASE, THYMINE | | Authors: | Walter, M.R, Cook, W.J, Cole, L.B, Short, S.A, Koszalka, G.W, Krenitsky, T.A, Ealick, S.E. | | Deposit date: | 1990-06-14 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of thymidine phosphorylase from Escherichia coli at 2.8 A resolution.
J.Biol.Chem., 265, 1990
|
|
1LYP
 
 | |
