6Z5L
 
 | |
6QWG
 
 | | Serial Femtosecond Crystallography Structure of Cu Nitrite Reductase from Achromobacter cycloclastes: Nitrite complex at Room Temperature | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Ebrahim, A.E, Moreno-Chicano, T, Appleby, M.V, Worrall, J.W, Duyvesteyn, H.M.E, Strange, R.W, Beale, J, Axford, D, Sherrell, D.A, Sugimoto, H, Owada, S, Tono, K, Owen, R.L. | | Deposit date: | 2019-03-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput structures of protein-ligand complexes at room temperature using serial femtosecond crystallography.
Iucrj, 6, 2019
|
|
6ZBY
 
 | |
1IT3
 
 | | Hagfish CO ligand hemoglobin | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin | | Authors: | Mito, M, Chong, K.T, Park, S.-Y, Tame, J.R. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of deoxy- and carbonmonoxyhemoglobin F1 from the hagfish Eptatretus burgeri
J.Biol.Chem., 277, 2002
|
|
5JYU
 
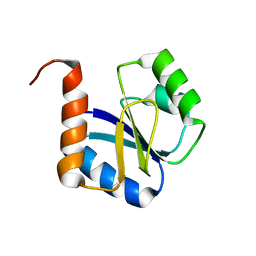 | |
6O30
 
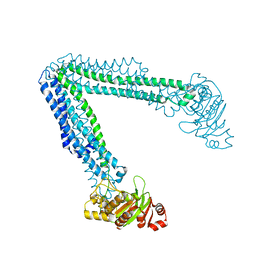 | | Lipid A transporter MsbA from Salmonella typhimurium | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Padayatti, P.S, Zhang, Q, Wilson, I.A, Lee, S.C, Stanfield, R.L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.47 Å) | | Cite: | Structural Insights into the Lipid A Transport Pathway in MsbA.
Structure, 27, 2019
|
|
1OIS
 
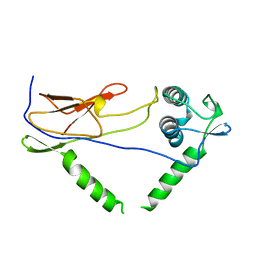 | | YEAST DNA TOPOISOMERASE I, N-TERMINAL FRAGMENT | | Descriptor: | DNA TOPOISOMERASE I | | Authors: | Lue, N, Sharma, A, Mondragon, A, Wang, J.C. | | Deposit date: | 1996-09-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 26 kDa yeast DNA topoisomerase I fragment: crystallographic structure and mechanistic implications.
Structure, 3, 1995
|
|
6ZS5
 
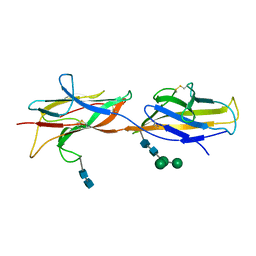 | | 3.5 A cryo-EM structure of human uromodulin filament core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stanisich, J.J, Zyla, D, Afanasyev, P, Xu, J, Pilhofer, M, Boeringer, D, Glockshuber, R. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism.
Elife, 9, 2020
|
|
7A46
 
 | | small conductance mechanosensitive channel YbiO | | Descriptor: | Putative transport protein | | Authors: | Flegler, V.J, Rasmussen, A, Rao, S, Wu, N, Zenobi, R, Sansom, M.S.P, Hedrich, R, Rasmussen, T, Boettcher, B. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The MscS-like channel YnaI has a gating mechanism based on flexible pore helices.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KWO
 
 | |
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
5KX2
 
 | |
5KWZ
 
 | |
7LAS
 
 | | Cryo-EM structure of PCV2 Replicase bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent helicase Rep, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*A)-3'), ... | | Authors: | Khayat, R. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Mbio, 12, 2021
|
|
5KX0
 
 | |
5KX1
 
 | |
5KWP
 
 | |
7LAR
 
 | | Cryo-EM structure of PCV2 Replicase bound to ssDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent helicase Rep, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Khayat, R. | | Deposit date: | 2021-01-06 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanism of DNA Interaction and Translocation by the Replicase of a Circular Rep-Encoding Single-Stranded DNA Virus.
Mbio, 12, 2021
|
|
1JM7
 
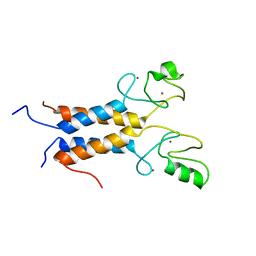 | | Solution structure of the BRCA1/BARD1 RING-domain heterodimer | | Descriptor: | BRCA1-ASSOCIATED RING DOMAIN PROTEIN 1, BREAST CANCER TYPE 1 SUSCEPTIBILITY PROTEIN, ZINC ION | | Authors: | Brzovic, P.S, Rajagopal, P, Hoyt, D.W, King, M.-C, Klevit, R.E. | | Deposit date: | 2001-07-17 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a BRCA1-BARD1 heterodimeric RING-RING complex.
Nat.Struct.Biol., 8, 2001
|
|
1XYR
 
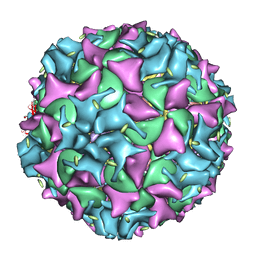 | | Poliovirus 135S cell entry intermediate | | Descriptor: | Genome polyprotein, Coat protein VP1, Coat protein VP2, ... | | Authors: | Bubeck, D, Filman, D.J, Cheng, N, Steven, A.C, Hogle, J.M, Belnap, D.M. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
J.Virol., 79, 2005
|
|
3FX2
 
 | |
1KSQ
 
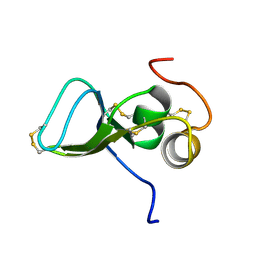 | | NMR Study of the Third TB Domain from Latent Transforming Growth Factor-beta Binding Protein-1 | | Descriptor: | LATENT TRANSFORMING GROWTH FACTOR BETA BINDING PROTEIN 1 | | Authors: | Lack, J, O'leary, J.M, Knott, V, Yuan, X, Rifkin, D.B, Handford, P.A, Downing, A.K. | | Deposit date: | 2002-01-14 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Third TB Domain from LTBP1 Provides Insight into Assembly
of the Large Latent Complex that Sequesters Latent TGF-beta.
J.Mol.Biol., 334, 2003
|
|
4M8T
 
 | |
1IT2
 
 | | Hagfish deoxy hemoglobin | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin | | Authors: | Mito, M, Chong, K.T, Park, S.-Y, Tame, J.R. | | Deposit date: | 2002-01-05 | | Release date: | 2002-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of deoxy- and carbonmonoxyhemoglobin F1 from the hagfish Eptatretus burgeri
J.Biol.Chem., 277, 2002
|
|
2Z4E
 
 | |
