7DGE
 
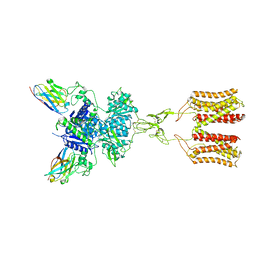 | | intermediate state of class C GPCR | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 1, nanobody | | Authors: | Zhang, J.Y, Wu, L.J, Luo, F, Hua, T, Liu, Z.J. | | Deposit date: | 2020-11-11 | | Release date: | 2021-09-22 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into the activation initiation of full-length mGlu1.
Protein Cell, 12, 2021
|
|
8TOT
 
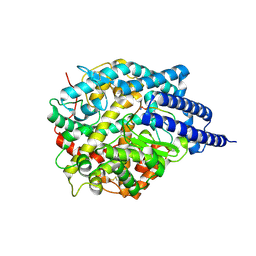 | | ACE2-peptide2 complex crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TA3
 
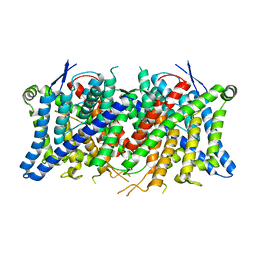 | | Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin | | Descriptor: | CHLORIDE ION, Chloride channel protein 2 | | Authors: | Xu, M, Neelands, T, Powers, A.S, Liu, Y, Miller, S, Pintilie, G, Du Bois, J, Dror, R.O, Chiu, W, Maduke, M. | | Deposit date: | 2023-06-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | CryoEM structures of the human CLC-2 voltage-gated chloride channel reveal a ball-and-chain gating mechanism.
Elife, 12, 2024
|
|
8TOU
 
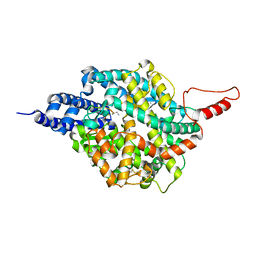 | | ACE2-peptide 2 complex crystal form 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Franck, C, Payne, R.J, Christie, M. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
2IRV
 
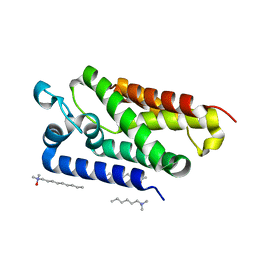 | | Crystal structure of GlpG, a rhomboid intramembrane serine protease | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, DODECYL-BETA-D-MALTOSIDE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Bibi, E, Fass, D, Ben-Shem, A. | | Deposit date: | 2006-10-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for intramembrane proteolysis by rhomboid serine proteases.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8UUA
 
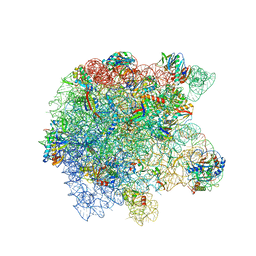 | | Cryo-EM structure of the Listeria innocua 50S ribosomal subunit in complex with HflXr (structure III) | | Descriptor: | 23S Ribosomal RNA, 5S Ribosomal RNA, GTPase HflX, ... | | Authors: | Seely, S.M, Basu, R.S, Gagnon, M.G. | | Deposit date: | 2023-10-31 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
2IC8
 
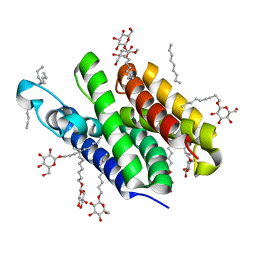 | | Crystal structure of GlpG | | Descriptor: | Protein glpG, nonyl beta-D-glucopyranoside | | Authors: | Ha, Y. | | Deposit date: | 2006-09-12 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a rhomboid family intramembrane protease.
Nature, 444, 2006
|
|
2J3F
 
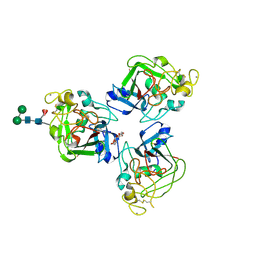 | | L-ficolin complexed to N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
8V01
 
 | |
2IDM
 
 | | 2.00 A Structure of T87I/Y106W Phosphono-CheY | | Descriptor: | ACETATE ION, Chemotaxis protein cheY | | Authors: | Halkides, C.J, Haas, R.M, McAdams, K.A, Casper, E.S, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structures of T87I phosphono-CheY and T87I/Y106W phosphono-CheY help to explain their binding affinities to the FliM and CheZ peptides.
Arch.Biochem.Biophys., 479, 2008
|
|
2J60
 
 | | H-ficolin complexed to D-fucose | | Descriptor: | ACETATE ION, CALCIUM ION, FICOLIN-3, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-09-21 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
8UWW
 
 | |
8UYD
 
 | |
2J95
 
 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5'-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-2,2'-BITHIOPHENE-5-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
2IWE
 
 | | Structure of a cavity mutant (H117G) of Pseudomonas aeruginosa azurin | | Descriptor: | 1,1'-HEXANE-1,6-DIYLBIS(1H-IMIDAZOLE), AZURIN, ZINC ION | | Authors: | De Jongh, T.E, Van Roon, A.M.M, Prudencio, M, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Click-Chemistry with an Active Site Variant of Azurin
Eur.J.Inorg.Chem., 2006, 2006
|
|
8UI0
 
 | |
7D6Y
 
 | | eIF4E in Complex with a Disulphide-Free Autonomous VH Domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Eukaryotic translation initiation factor 4E, VH Domain (1C5), ... | | Authors: | Brown, C.J, Frosi, Y, Jiang, S, Lin, Y.C. | | Deposit date: | 2020-10-02 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.668 Å) | | Cite: | Engineering Disulphide-Free Autonomous Antibody VH Domains to modulate intracellular pathways
To Be Published
|
|
8S86
 
 | | human PLD3 homodimer structure | | Descriptor: | 5'-3' exonuclease PLD3 | | Authors: | Lammens, K. | | Deposit date: | 2024-03-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Lysosomal endonuclease RNase T2 and PLD exonucleases cooperatively generate RNA ligands for TLR7 activation.
Immunity, 57, 2024
|
|
2IJ5
 
 | |
7F94
 
 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels | | Descriptor: | A C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
2IMF
 
 | | 2-Hydroxychromene-2-carboxylate Isomerase: a Kappa Class Glutathione-S-Transferase from Pseudomonas putida | | Descriptor: | 2-hydroxychromene-2-carboxylate isomerase, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, 4-(2-METHOXYPHENYL)-2-OXOBUT-3-ENOIC ACID, ... | | Authors: | Thompson, L.C, Ladner, J.E, Codreanu, S.G, Harp, J, Gilliland, G.L, Armstrong, R.N. | | Deposit date: | 2006-10-04 | | Release date: | 2007-06-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 2-Hydroxychromene-2-carboxylic acid isomerase: a kappa class glutathione transferase from Pseudomonas putida.
Biochemistry, 46, 2007
|
|
2INZ
 
 | | Crystal Structure of Aldose Reductase complexed with 2-Hydroxyphenylacetic Acid | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Carlson, E, Brownlee, J.M. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
8QNS
 
 | |
2I1J
 
 | | Moesin from Spodoptera frugiperda at 2.1 angstroms resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Moesin, ... | | Authors: | Li, Q, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2006-08-14 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Self-masking in an Intact ERM-merlin Protein: An Active Role for the Central alpha-Helical Domain.
J.Mol.Biol., 365, 2007
|
|
2HZL
 
 | | Crystal structures of a sodium-alpha-keto acid binding subunit from a TRAP transporter in its closed forms | | Descriptor: | PYRUVIC ACID, SODIUM ION, TRAP-T family sorbitol/mannitol transporter, ... | | Authors: | Gonin, S, Arnoux, P, Pierru, B, Alonso, B, Sabaty, M, Pignol, D. | | Deposit date: | 2006-08-09 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding.
Bmc Struct.Biol., 7, 2007
|
|
