4V93
 
 | | Fitted coordinates for Lumbricus terrestris hemoglobin cryo-EM complex (EMD-2627) | | Descriptor: | EXTRACELLULAR GLOBIN-2, EXTRACELLULAR GLOBIN-3, EXTRACELLULAR GLOBIN-4, ... | | Authors: | Chen, W.T, Chen, Y.C, Liou, H.H, Chao, C.Y. | | Deposit date: | 2014-04-16 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural Basis for Cooperative Oxygen Binding and Bracelet-Assisted Assembly of Lumbricus Terrestris Hemoglobin
To be Published
|
|
4U1R
 
 | |
4URA
 
 | | Crystal structure of human JMJD2A in complex with compound 14a | | Descriptor: | 1,2-ETHANEDIOL, 2-(2H-1,2,3-triazol-4-yl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Krojer, T, England, K.S, Vollmar, M, Crawley, L, Williams, E, Riesebos, E, Szykowska, A, Burgess-Brown, N, Oppermann, U, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor.
Medchemcomm, 5, 2014
|
|
4UZC
 
 | |
4U9Q
 
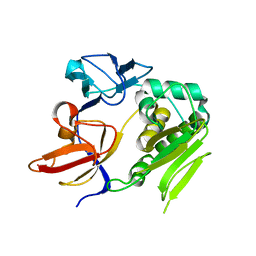 | | Crystal structure of NqrA in spacegroup P21 | | Descriptor: | Na(+)-translocating NADH-quinone reductase subunit A | | Authors: | Fritz, G. | | Deposit date: | 2014-08-06 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the V. cholerae Na+-pumping NADH:quinone oxidoreductase.
Nature, 516, 2014
|
|
4UU1
 
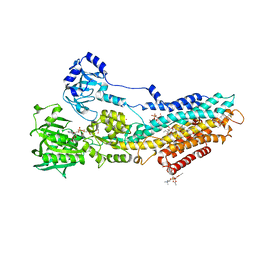 | | CRYSTAL STRUCTURE OF (SR) CALCIUM-ATPASE E2(TG) IN THE PRESENCE OF DOPC | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Drachmann, N.D, Olesen, C, Moeller, J.V, Guo, Z, Nissen, P, Bublitz, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparing Crystal Structures of Ca(2+) -ATPase in the Presence of Different Lipids.
FEBS J., 281, 2014
|
|
4UWP
 
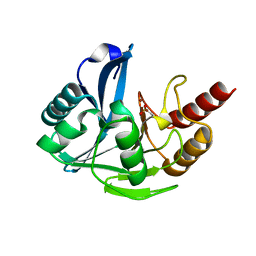 | | Penta Zn1 coordination. Leu224 in VIM-26 from Klebsiella pneumoniae has implications for drug binding. | | Descriptor: | METALLO-BETA-LACTAMASE VIM-26, ZINC ION | | Authors: | Leiros, H.-K.S, Edvardsen, K.S.W, Bjerga, G.E.K, Samuelsen, O. | | Deposit date: | 2014-08-14 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Characterization of Vim-26 Show that Leu224 Has Implications for the Substrate Specificity of Vim Metallo-Beta-Lactamases.
FEBS J., 282, 2015
|
|
4UX8
 
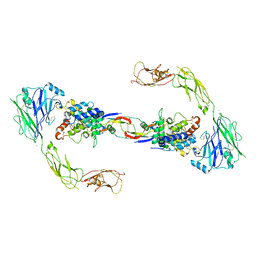 | | RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF FAMILY RECEPTOR ALPHA-1, ... | | Authors: | Goodman, K, Kjaer, S, Beuron, F, Knowles, P, Nawrotek, A, Burns, E, Purkiss, A, George, R, Santoro, M, Morris, E.P, McDonald, N.Q. | | Deposit date: | 2014-08-19 | | Release date: | 2014-10-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Ret Recognition of Gdnf-Gfralpha1 Ligand by a Composite Binding Site Promotes Membrane-Proximal Self-Association.
Cell Rep., 8, 2014
|
|
5OW9
 
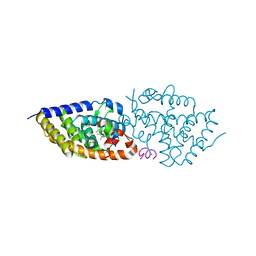 | | Vitamin D receptor complex | | Descriptor: | (1~{S},3~{Z})-3-[(2~{E})-2-[(1~{S},3~{a}~{S},7~{a}~{S})-7~{a}-methyl-1-[(2~{S})-6-methyl-2-oxidanyl-heptan-2-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexan-1-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Li, W. | | Deposit date: | 2017-08-31 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Investigation of 20S-hydroxyvitamin D3 analogs and their 1 alpha-OH derivatives as potent vitamin D receptor agonists with anti-inflammatory activities.
Sci Rep, 8, 2018
|
|
4V34
 
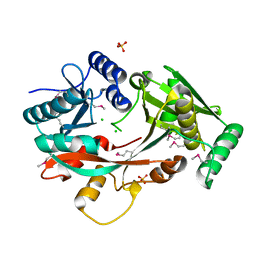 | | The Structure of A-PGS from Pseudomonas aeruginosa (SeMet derivative) | | Descriptor: | ALANYL-TRNA-DEPENDENT L-ALANYL- PHOPHATIDYLGLYCEROL SYNTHASE, CHLORIDE ION, SULFATE ION | | Authors: | Krausze, J, Hebecker, S, Hasenkampf, T, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UBS
 
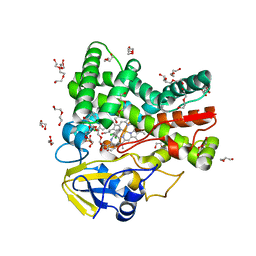 | | The crystal structure of cytochrome P450 105D7 from Streptomyces avermitilis in complex with Diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Xu, L.H, Ikeda, H, Arakawa, T, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2014-08-13 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the 4'-hydroxylation of diclofenac by a microbial cytochrome P450 monooxygenase.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
4V86
 
 | |
4V8L
 
 | |
4UQX
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
J.Biol.Chem., 289, 2014
|
|
4UT5
 
 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa strain PA7 in complex with lewis a tetrasaccharide | | Descriptor: | CALCIUM ION, LECB LECTIN, beta-D-galactopyranose, ... | | Authors: | Boukerb, A.M, Decor, A, Tabaroni, R, Varrot, A, Debentzmann, S, Vidal, S, Imberty, A, Cournoyer, B. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genomic Rearrangements and Functional Diversification of Leca and Lecb Lectin-Coding Regions Impacting the Efficacy of Glycomimetics Directed Against Pseudomonas Aeruginosa.
Front.Microbiol., 7, 2016
|
|
4UYT
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo11 from Saccharomyces cerevisiae | | Descriptor: | ACETATE ION, FLOCCULATION PROTEIN FLO11, SODIUM ION | | Authors: | Kraushaar, T, Veelders, M, Brueckner, S, Rhinow, D, Moesch, H.U, Essen, L.O. | | Deposit date: | 2014-09-03 | | Release date: | 2015-08-12 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Interactions by the Fungal Flo11 Adhesin Depend on a Fibronectin Type III-Like Adhesin Domain Girdled by Aromatic Bands.
Structure, 23, 2015
|
|
4V4F
 
 | | The structure of the trp RNA-binding attenuation protein (TRAP) bound to a RNA molecule containing UAGAU repeats | | Descriptor: | 5'-R(*UP*AP*GP*AP*UP)-3', TRANSCRIPTION ATTENUATION PROTEIN MTRB, TRYPTOPHAN | | Authors: | Hopcroft, N.H, Manfredo, A, Wendt, A.L, Brzozowski, A.M, Gollnick, P, Antson, A.A. | | Deposit date: | 2003-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Interaction of RNA with Trap: The Role of Triplet Repeats and Separating Spacer Nucleotides
J.Mol.Biol., 338, 2004
|
|
4U39
 
 | | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis | | Descriptor: | Cell division factor, Cell division protein FtsZ, PHOSPHATE ION | | Authors: | Bisson-Filho, A.W, Discola, K.F, Castellen, P, Blasios, V, Martins, A, Sforca, M.L, Garcia, W, Zeri, A.C, Erickson, H.P, Dessen, A, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis
To be Published
|
|
4WA3
 
 | | The crystal structure of neuraminidase from a H3N8 influenza virus isolated from New England harbor seals | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NICKEL (II) ION, ... | | Authors: | Yang, H, Villanueva, J.M, Gubareva, L.V, Stevens, J. | | Deposit date: | 2014-08-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural and Functional Analysis of Surface Proteins from an A(H3N8) Influenza Virus Isolated from New England Harbor Seals.
J.Virol., 89, 2015
|
|
4WBO
 
 | |
4UPU
 
 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
4USU
 
 | | Crystal structure of human soluble Adenylyl Cyclase in complex with alpha,beta-methyleneadenosine-5'-triphosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE CYCLASE TYPE 10, CALCIUM ION, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2014-07-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Analysis of Human Soluble Adenylyl Cyclase and Crystal Structures of its Nucleotide Complexes -Implications for Cyclase Catalysis and Evolution.
FEBS J., 281, 2014
|
|
4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
4U64
 
 | | Structure of the periplasmic output domain of the Legionella pneumophila LapD ortholog CdgS9 in the apo state | | Descriptor: | Two component histidine kinase, GGDEF domain protein/EAL domain protein | | Authors: | Chatterjee, D, Cooley, R.B, Boyd, C.D, Mehl, R.A, O'Toole, G.A, Sondermann, H.S. | | Deposit date: | 2014-07-27 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP.
Elife, 3, 2014
|
|
4UZV
 
 | | Structure of a triple mutant of ASV-TfTrHb | | Descriptor: | ACETATE ION, HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Baiocco, P, Bonamore, A, Sciamanna, N, Ilari, A, Boechi, L, Boffi, A, Smulevich, G, Feis, A. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Role of Local Structure and Dynamics of Small Ligand Migration in Proteins: A Study of a Mutated Truncated Hemoprotein from Thermobifida Fusca by Time Resolved Mir Spectroscopy.
J.Phys.Chem.B, 118, 2014
|
|
