5L1Q
 
 | | X-ray Structure of Cytochrome P450 PntM with Dihydropentalenolactone F | | Descriptor: | Dihydropentalenolactone F, PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5L1S
 
 | |
3UHE
 
 | | HBI (M37V,L73I) CO bound | | Descriptor: | CARBON MONOXIDE, Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UHS
 
 | | HBI (L36M) deoxy | | Descriptor: | Globin-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ren, Z, Srajer, V, Knapp, J.E, Royer Jr, W.E. | | Deposit date: | 2011-11-03 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative macromolecular device revealed by meta-analysis of static and time-resolved structures.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1T36
 
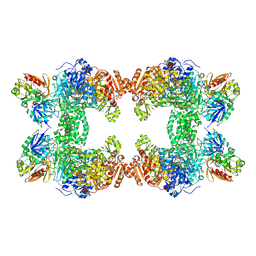 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
2IS9
 
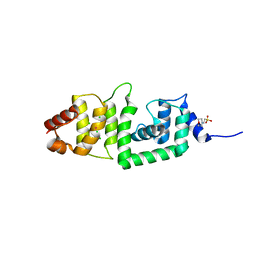 | | Structure of yeast DCN-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | Authors: | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | Deposit date: | 2006-10-16 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
1N75
 
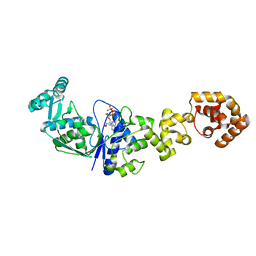 | | Crystal structure of Thermus thermophilus glutamyl-tRNA synthetase complexed with ATP. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glutamyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sekine, S, Nureki, O, Dubois, D.Y, Bernier, S, Chenevert, R, Lapointe, J, Vassylyev, D.G, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-11-12 | | Release date: | 2003-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding
EMBO J., 22, 2003
|
|
2J4C
 
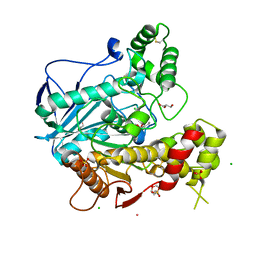 | | Structure of human Butyrylcholinesterase in complex with 10mM HgCl2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Colletier, J.P, Frasco, M.F, Carvalho, F, Guilhermino, L, Stojan, J, Fournier, D, Weik, M. | | Deposit date: | 2006-08-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanisms of Cholinesterase Inhibition by Inorganic Mercury.
FEBS J., 274, 2007
|
|
4G1D
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{[3,4-bis(hydroxymethyl)benzyl]oxy}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-10 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
1F1H
 
 | |
2A91
 
 | | Crystal structure of ErbB2 domains 1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Garrett, T.P.J, McKern, N.M, Lou, M, Elleman, T.C, Adams, T.E, Lovrecz, G.O, Kofler, M, Jorissen, R.N, Nice, E.C, Burgess, A.W. | | Deposit date: | 2005-07-11 | | Release date: | 2005-07-26 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of a Truncated ErbB2 Ectodomain Reveals an Active Conformation, Poised to Interact with Other ErbB Receptors
Mol.Cell, 11, 2003
|
|
2H5A
 
 | | Complex of the enzyme PMM/PGM with xylose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-xylopyranose, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Regni, C, Shackelford, G.S, Beamer, L.J. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Complexes of the enzyme phosphomannomutase/phosphoglucomutase with a slow substrate and an inhibitor.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3NSP
 
 | | Crystal structure of tetrameric RXRalpha-LBD | | Descriptor: | Retinoid X receptor, alpha | | Authors: | Zhang, H, Hu, T, Li, L, Zhou, R, Chen, L, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Danthron functions as a retinoic X receptor antagonist by stabilizing tetramers of the receptor.
J.Biol.Chem., 286, 2011
|
|
4G20
 
 | | Structural basis for the accommodation of bis- and tris-aromatic derivatives in Vitamin D Nuclear Receptor | | Descriptor: | 3-(5'-{2-[3,4-bis(hydroxymethyl)phenyl]ethyl}-2'-methyl-2-propylbiphenyl-4-yl)pentan-3-ol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Ciesielski, F, Sato, Y, Moras, D, Rochel, N. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the accommodation of bis- and tris-aromatic derivatives in vitamin d nuclear receptor.
J.Med.Chem., 55, 2012
|
|
3FI0
 
 | | Crystal Structure Analysis of B. stearothermophilus Tryptophanyl-tRNA Synthetase Complexed with Tryptophan, AMP, and Inorganic Phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, TRYPTOPHAN, ... | | Authors: | Laowanapiban, P, Kapustina, M, Vonrhein, C, Delarue, M, Koehl, P, Carter Jr, C.W. | | Deposit date: | 2008-12-10 | | Release date: | 2009-02-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Independent saturation of three TrpRS subsites generates a partially assembled state similar to those observed in molecular simulations.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
3SX7
 
 | | Crystal structure of ABBA+UDP+Gal with Glycerol as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Histo-blood group ABO system transferase, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SXA
 
 | | Crystal structure of ABBB+UDP+Gal with Glycerol as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Histo-blood group ABO system transferase, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SXG
 
 | | Crystal structure of AAAA+UDP+Gal with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SX5
 
 | | Crystal structure of AABB+UDP+Gal with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SXB
 
 | | Crystal structure of ABBB+UDP+Gal with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3SXC
 
 | | Crystal structure of BBBB+UDP+Gal with Glycerol as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
9B5B
 
 | |
5OMW
 
 | |
9B5A
 
 | |
9B59
 
 | |
