7VFH
 
 | |
2J08
 
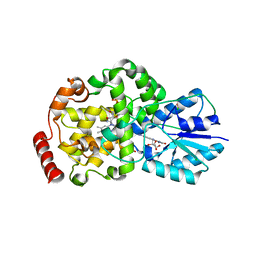 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
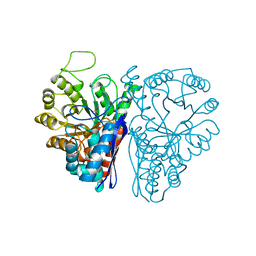
3ENL
 
 | |
4EP2
 
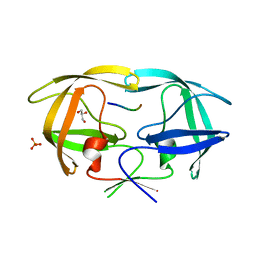 | | Crystal Structure of inactive single chain wild-type HIV-1 Protease in Complex with the substrate RT-RH | | Descriptor: | GLYCEROL, PHOSPHATE ION, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
4EEO
 
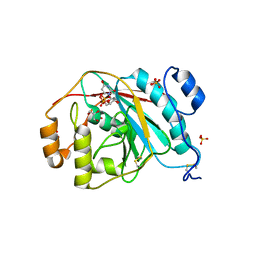 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-GlcNAc-ALPHA-benzyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-benzyl 2-acetamido-2-deoxy-alpha-D-glucopyranoside, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EEM
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-MAN-ALPHA-methyl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-methyl alpha-D-mannopyranoside, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
4EPJ
 
 | | Crystal Structure of inactive single chain wild-type HIV-1 Protease in Complex with the substrate p2-NC | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
4KVO
 
 | |
1F2J
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ALDOLASE FROM T. BRUCEI | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE, GLYCOSOMAL | | Authors: | Chudzik, D.M, Michels, P.A, De Walque, S, Hol, W.G.J. | | Deposit date: | 2000-05-25 | | Release date: | 2000-07-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of type 2 peroxisomal targeting signals in two trypanosomatid aldolases.
J.Mol.Biol., 300, 2000
|
|
3E76
 
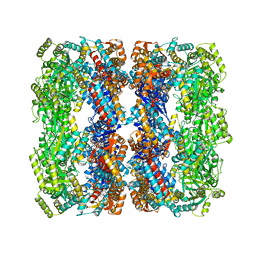 | | Crystal structure of Wild-type GroEL with bound Thallium ions | | Descriptor: | 60 kDa chaperonin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kiser, P.D, Lorimer, G.H, Palczewski, K. | | Deposit date: | 2008-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Use of thallium to identify monovalent cation binding sites in GroEL.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7VKF
 
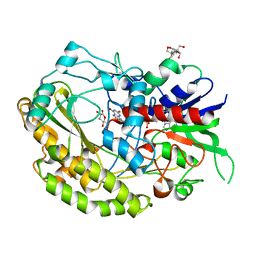 | | Reduced enzyme of FAD-dpendent Glucose Dehydrogenase complex with D-glucono-1,5-lactone at pH8.5 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-glucono-1,5-lactone, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Nakajima, Y, Nishiya, Y, Ito, K. | | Deposit date: | 2021-09-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational change of catalytic residue in reduced enzyme of FAD-dependent Glucose Dehydrogenase at pH6.5
To Be Published
|
|
3W6D
 
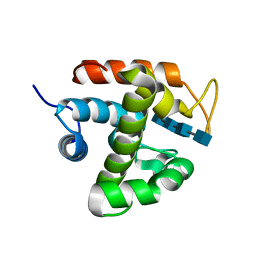 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
1RFF
 
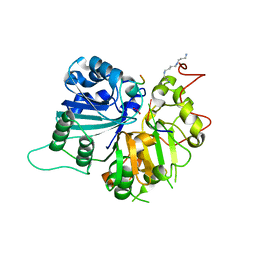 | | Crystal structure of human Tyrosyl-DNA Phosphodiesterase complexed with vanadate, octapeptide KLNYYDPR, and tetranucleotide AGTT. | | Descriptor: | 5'-D(*AP*GP*TP*T)-3', SPERMINE, Topoisomerase I-Derived Peptide, ... | | Authors: | Davies, D.R, Interthal, H, Champoux, J.J, Hol, W.G. | | Deposit date: | 2003-11-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Explorations of peptide and oligonucleotide binding sites of tyrosyl-DNA phosphodiesterase using vanadate complexes.
J.Med.Chem., 47, 2004
|
|
7VFR
 
 | | GltA N83K mutant from Bifidobacterium infantis JCM 1222 complexed with lacto-N-tetraose | | Descriptor: | Extracellular solute-binding protein, family 1, SULFATE ION, ... | | Authors: | Sato, M, Sakanaka, M, Katayama, T, Fushinobu, S. | | Deposit date: | 2021-09-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Short stretch sequence of a milk oligosaccharide transporter represents the phenotypic selection trajectory in infant gut microbiome
To Be Published
|
|
2BKZ
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-404611 | | Descriptor: | 1-[4-(AMINOSULFONYL)PHENYL]-1,6-DIHYDROPYRAZOLO[3,4-E]INDAZOLE-3-CARBOXAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | DAlessio, R, Bargiottia, A, Metz, S, Brasca, M.G, Cameron, A, Ermoli, A, Marsiglio, A, Polucci, P, Roletto, F, Tibolla, M, Vazquez, M.L, Vulpetti, A, Pevarello, P. | | Deposit date: | 2005-02-23 | | Release date: | 2006-03-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Benzodipyrazoles: A New Class of Potent Cdk2 Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7VFQ
 
 | | Wild type GltA from Bifidobacterium infantis JCM 1222 complexed with lacto-N-tetraose | | Descriptor: | Extracellular solute-binding protein, family 1, SULFATE ION, ... | | Authors: | Sato, M, Sakanaka, M, Katayama, T, Fushinobu, S. | | Deposit date: | 2021-09-13 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Short stretch sequence of a milk oligosaccharide transporter represents the phenotypic selection trajectory in infant gut microbiome
To Be Published
|
|
4EQJ
 
 | | Crystal Structure of inactive single chain variant of HIV-1 Protease in Complex with the substrate RT-RH | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
1ORM
 
 | |
2C16
 
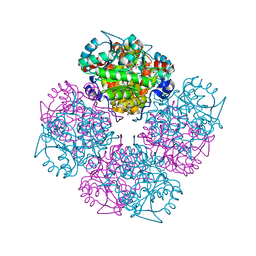 | | 5-(4-Carboxy-2-oxo-butane-1-sulfinyl)-4-oxo-pentanoic acid acid bound to Porphobilinogen synthase from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Frere, F, Nentwich, M, Gacond, S, Heinz, D.W, Neier, R, Frankenberg-Dinkel, N. | | Deposit date: | 2005-09-11 | | Release date: | 2006-06-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Probing the Active Site of Pseudomonas Aeruginosa Porphobilinogen Synthase Using Newly Developed Inhibitors.
Biochemistry, 45, 2006
|
|
1P6W
 
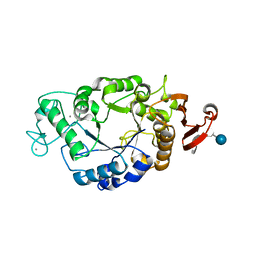 | | Crystal structure of barley alpha-amylase isozyme 1 (AMY1) in complex with the substrate analogue, methyl 4I,4II,4III-tri-thiomaltotetraoside (thio-DP4) | | Descriptor: | CALCIUM ION, PROTEIN (Alpha-amylase type A isozyme), alpha-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of barley alpha-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs
Structure, 11, 2003
|
|
1FMZ
 
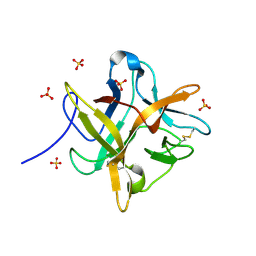 | | CRYSTAL STRUCTURE OF A MUTANT WINGED BEAN CHYMOTRYPSIN INHIBITOR PROTEIN, N14K. | | Descriptor: | CHYMOTRYPSIN INHIBITOR 3, SULFATE ION | | Authors: | Dattagupta, J.K, Chakrabarti, C, Ravichandran, S, Dasgupta, J, Ghosh, S. | | Deposit date: | 2000-08-19 | | Release date: | 2001-02-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of Asn14 in the stability and conformation of the reactive-site loop of winged bean chymotrypsin inhibitor: crystal structures of two point mutants Asn14-->Lys and Asn14-->Asp.
PROTEIN ENG., 14, 2001
|
|
1FD7
 
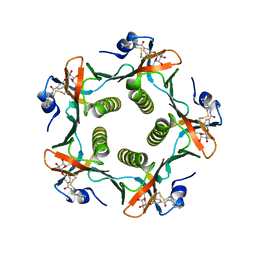 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER WITH BOUND LIGAND BMSC001 | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, N-BENZYL-3-(ALPHA-D-GALACTOS-1-YL)-BENZAMIDE | | Authors: | Fan, E, Merritt, E.A, Pickens, J, Ahn, M, Hol, W.G.J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploration of the GM1 receptor-binding site of heat-labile enterotoxin and cholera toxin by phenyl-ring-containing galactose derivatives.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4CSU
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with ObgE | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | Feng, B, Mandava, C.S, Guo, Q, Wang, J, Cao, W, Li, N, Zhang, Y, Zhang, Y, Wang, Z, Wu, J, Sanyal, S, Lei, J, Gao, N. | | Deposit date: | 2014-03-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural and Functional Insights Into the Mode of Action of a Universally Conserved Obg Gtpase.
Plos Biol., 12, 2014
|
|
5TMN
 
 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
4IDP
 
 | | human atlastin-1 1-446, N440T, GppNHp | | Descriptor: | Atlastin-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Byrnes, L.J, Singh, A, Szeto, K, Benvin, N.M, O'Donnell, J.P, Zipfel, W.R, Sondermann, H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural basis for conformational switching and GTP loading of the large G protein atlastin.
Embo J., 32, 2013
|
|
