3SEV
 
 | | Zn-mediated Trimer of Maltose-binding Protein E310H/K314H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, Maltose-binding periplasmic protein, ZINC ION, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3JVT
 
 | | Calcium-bound Scallop Myosin Regulatory Domain (Lever Arm) with Reconstituted Complete Light Chains | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Himmel, D.M, Mui, S, O'Neall-Hennessey, E, Szent-Gyorgyi, A, Cohen, C. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The on-off switch in regulated myosins: different triggers but related mechanisms.
J.Mol.Biol., 394, 2009
|
|
3JX8
 
 | |
3JZ7
 
 | |
3S94
 
 | | Crystal structure of LRP6-E1E2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3K11
 
 | |
3K1T
 
 | |
3K23
 
 | | Glucocorticoid Receptor with Bound D-prolinamide 11 | | Descriptor: | 1-{[3-(4-{[(2R)-4-(5-fluoro-2-methoxyphenyl)-2-hydroxy-4-methyl-2-(trifluoromethyl)pentyl]amino}-6-methyl-1H-indazol-1-yl)phenyl]carbonyl}-D-prolinamide, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3S8Z
 
 | | Crystal structure of LRP6-E3E4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6 | | Authors: | Cheng, Z, Xu, W. | | Deposit date: | 2011-05-31 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the extracellular domain of LRP6 and its complex with DKK1.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3SER
 
 | | Zn-mediated Polymer of Maltose-binding Protein K26H/K30H by Synthetic Symmetrization | | Descriptor: | CALCIUM ION, CHLORIDE ION, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3S6Z
 
 | |
3SEY
 
 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SEU
 
 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form III) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
7MNI
 
 | | Crystal structure of the N-terminal domain of NUP88 in complex with NUP98 C-terminal Autoproteolytic Domain | | Descriptor: | Nuclear pore complex protein Nup88, Nuclear pore complex protein Nup98 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNJ
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
3K2U
 
 | | Crystal structure of HGFA in complex with the allosteric inhibitory antibody Fab40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody, Fab fragment, ... | | Authors: | Ganesan, R, Eigenbrot, C, Shia, S. | | Deposit date: | 2009-09-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unraveling the allosteric mechanism of serine protease inhibition by an antibody.
Structure, 17, 2009
|
|
7MNO
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) I656V mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.73 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNK
 
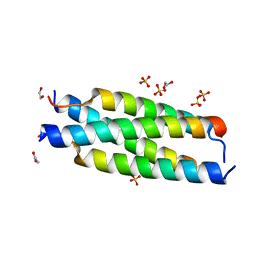 | | Crystal structure of the tetramerization element of NUP358/RanBP2 (residues 805-832) | | Descriptor: | 1,2-ETHANEDIOL, E3 SUMO-protein ligase RanBP2, SULFATE ION | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNM
 
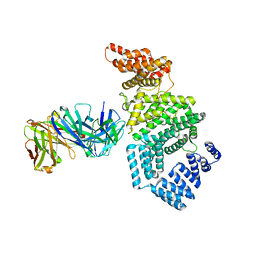 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) T585M mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNL
 
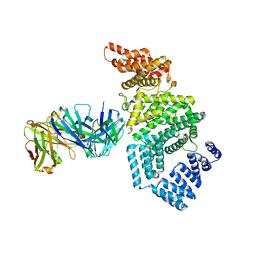 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNN
 
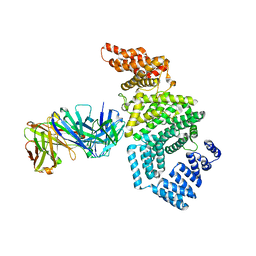 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) T653I mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7N60
 
 | | The crystal structure of 4-methoxybenzoate-bound CYP199A S244D mutant | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lau, I.C.K, Coleman, T, Bell, S.G, Bruning, J.B. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | The crystal structure of 4-methoxybenzoate-bound CYP199A S244D mutant
To Be Published
|
|
7N8T
 
 | | Crystal Structure of AMP-bound Human JNK2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, HEXAETHYLENE GLYCOL, Mitogen-activated protein kinase 9 | | Authors: | Li, L, Gurbani, D, Westover, K.D. | | Deposit date: | 2021-06-15 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of a Covalent Inhibitor of c-Jun N-Terminal Protein Kinase (JNK) 2/3 with Selectivity over JNK1.
J.Med.Chem., 66, 2023
|
|
7N9H
 
 | |
