1R5I
 
 | | Crystal structure of the MAM-MHC complex | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Zhao, Y, Li, Z, Drozd, S.J, Guo, Y, Mourad, W, Li, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen-MHC complex.
Structure, 12, 2004
|
|
1R5J
 
 | | Crystal Structure of a Phosphotransacetylase from Streptococcus pyogenes | | Descriptor: | putative phosphotransacetylase | | Authors: | Xu, Q.S, Shin, D.H, Pufan, R, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-10-10 | | Release date: | 2004-04-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a phosphotransacetylase from Streptococcus pyogenes.
Proteins, 55, 2004
|
|
1R5K
 
 | | Human Estrogen Receptor alpha Ligand-Binding Domain In Complex With GW5638 | | Descriptor: | (2E)-3-{4-[(1E)-1,2-DIPHENYLBUT-1-ENYL]PHENYL}ACRYLIC ACID, Estrogen receptor | | Authors: | Wu, Y.-L, Yang, X, Ren, Z, McDonnell, D.P, Norris, J.D, Willson, T.M, Greene, G.L. | | Deposit date: | 2003-10-10 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for an unexpected mode of SERM-mediated ER antagonism.
Mol.Cell, 18, 2005
|
|
1R5L
 
 | | Crystal Structure of Human Alpha-Tocopherol Transfer Protein Bound to its Ligand | | Descriptor: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, PROTEIN (Alpha-tocopherol transfer protein) | | Authors: | Min, K.C, Kovall, R.A, Hendrickson, W.A. | | Deposit date: | 2003-10-10 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human alpha-tocopherol transfer protein bound to its ligand: Implications for ataxia with vitamin E deficiency
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1R5M
 
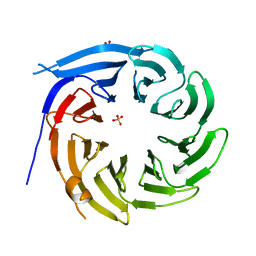 | |
1R5N
 
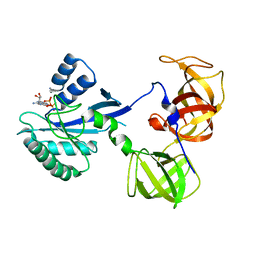 | | Crystal Structure Analysis of sup35 complexed with GDP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5O
 
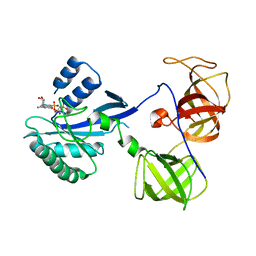 | | crystal structure analysis of sup35 complexed with GMPPNP | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kong, C, Song, H. | | Deposit date: | 2003-10-11 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5P
 
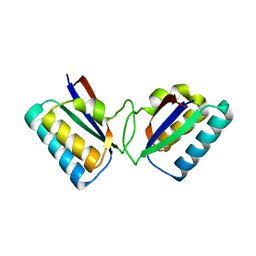 | |
1R5Q
 
 | |
1R5S
 
 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
1R5T
 
 | | The Crystal Structure of Cytidine Deaminase CDD1, an Orphan C to U editase from Yeast | | Descriptor: | Cytidine deaminase, ZINC ION | | Authors: | Xie, K, Sowden, M.P, Dance, G.S.C, Torelli, A.T, Smith, H.C, Wedekind, J.E. | | Deposit date: | 2003-10-13 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R5U
 
 | | RNA POLYMERASE II TFIIB COMPLEX | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | Authors: | Bushnell, D.A, Westover, K.D, Davis, R, Kornberg, R.D. | | Deposit date: | 2003-10-13 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms.
Science, 303, 2004
|
|
1R5V
 
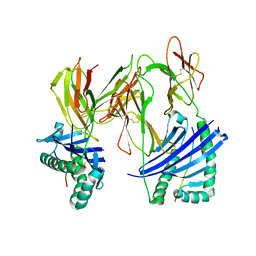 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E.J, He, X.L, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-02-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation.
Mol.Cell, 12, 2003
|
|
1R5W
 
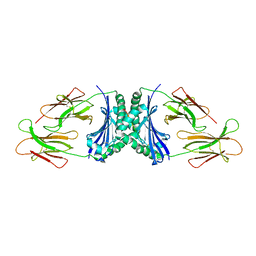 | | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T-cell activation | | Descriptor: | H-2 class II histocompatibility antigen, E-K alpha chain, MHC H2-IE-beta, ... | | Authors: | Krogsgaard, M, Prado, N, Adams, E, He, X, Chow, D.C, Wilson, D.B, Garcia, K.C, Davis, M.M. | | Deposit date: | 2003-10-13 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation
Mol.Cell, 12, 2003
|
|
1R5X
 
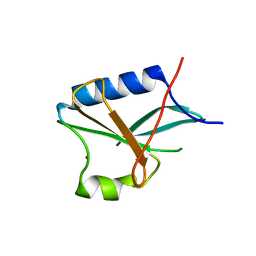 | |
1R5Y
 
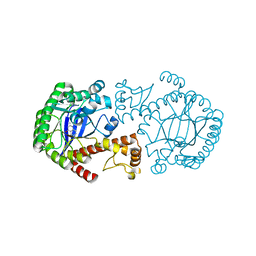 | | Crystal Structure of TGT in complex with 2,6-Diamino-3H-Quinazolin-4-one Crystallized at PH 5.5 | | Descriptor: | 2,6-DIAMINO-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Garcia, G.A, Stubbs, M.T, Klebe, G. | | Deposit date: | 2003-10-13 | | Release date: | 2004-04-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1R5Z
 
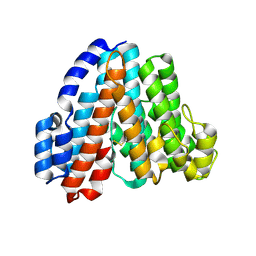 | | Crystal Structure of Subunit C of V-ATPase | | Descriptor: | V-type ATP synthase subunit C | | Authors: | Iwata, M, Imamura, H, Stambouli, E, Ikeda, C, Tamakoshi, M, Nagata, K, Makyio, H, Hankamer, B, Barber, J, Yoshida, M, Yokoyama, K, Iwata, S. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1R61
 
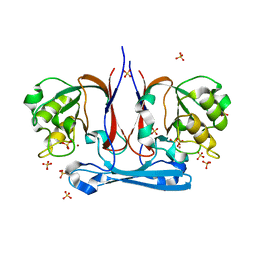 | | The structure of predicted metal-dependent hydrolase from Bacillus stearothermophilus | | Descriptor: | SULFATE ION, ZINC ION, metal-dependent hydrolase | | Authors: | Maderova, J, Borek, D, Tomchick, D, Joachimiak, A, Collart, F, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of potential metal-dependent hydrolase with cyclase activity
To be Published
|
|
1R62
 
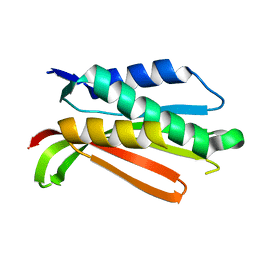 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | Descriptor: | Nitrogen regulation protein NR(II) | | Authors: | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | Deposit date: | 2003-10-14 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1R64
 
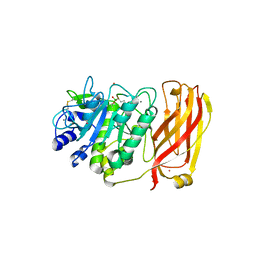 | | The 2.2 A crystal structure of Kex2 protease in complex with Ac-Arg-Glu-Lys-boroArg peptidyl boronic acid inhibitor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Holyoak, T, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Differences in Substrate Selectivity in Kex2 and Furin Protein Convertases
Biochemistry, 43, 2004
|
|
1R65
 
 | | Crystal structure of ferrous soaked Ribonucleotide Reductase R2 subunit (wildtype) at pH 5 from E. coli | | Descriptor: | FE (II) ION, MERCURY (II) ION, Ribonucleoside-diphosphate reductase 1 beta chain | | Authors: | Voegtli, W.C, Sommerhalter, M, Saleh, L, Baldwin, J, Bollinger Jr, J.M, Rosenzweig, A.C. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Variable coordination geometries at the diiron(II) active site of ribonucleotide reductase R2.
J.Am.Chem.Soc., 125, 2003
|
|
1R66
 
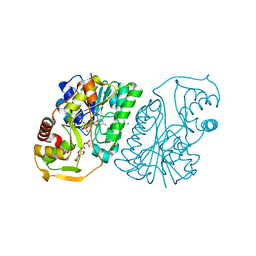 | | Crystal Structure of DesIV (dTDP-glucose 4,6-dehydratase) from Streptomyces venezuelae with NAD and TYD bound | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TDP-glucose-4,6-dehydratase, ... | | Authors: | Allard, S.T.M, Cleland, W.W, Holden, H.M. | | Deposit date: | 2003-10-14 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High Resolution X-ray Structure of dTDP-Glucose 4,6-Dehydratase from Streptomyces venezuelae
J.Biol.Chem., 279, 2004
|
|
1R67
 
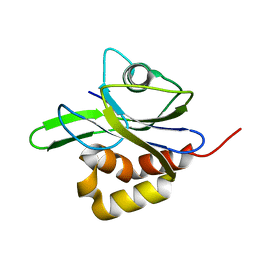 | | Y104A MUTANT OF E.COLI IPP ISOMERASE | | Descriptor: | Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION | | Authors: | Wouters, J. | | Deposit date: | 2003-10-15 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural role for Tyr-104 in Escherichia coli isopentenyl-diphosphate isomerase: site-directed mutagenesis, enzymology, and protein crystallography.
J.Biol.Chem., 281, 2006
|
|
1R68
 
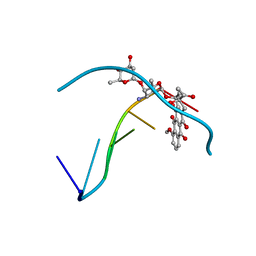 | | Role of the amino sugar in DNA binding of disaccharide anthracyclines: crystal structure of MAR70/d(CGATCG) complex | | Descriptor: | 4'-EPI-4'-(2-DEOXYFUCOSE)DAUNOMYCIN, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Temperini, C, Cirilli, M, Aschi, M, Ughetto, G. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Role of the amino sugar in DNA binding of disaccharide anthracyclines: crystal structure of the complex MAR70/d(CGATCG).
BIOORG.MED.CHEM., 13, 2005
|
|
