7K56
 
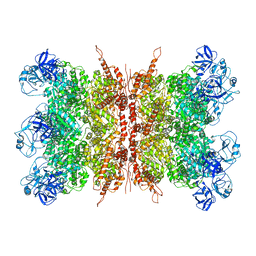 | | Structure of VCP dodecamer purified from H1299 cells | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
2GDS
 
 | |
7K57
 
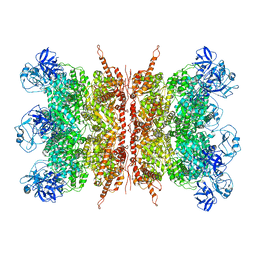 | | Structure of apo VCP dodecamer generated from bacterially recombinant VCP/p97 | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Yu, G, Bai, Y, Li, K, Jiang, W, Zhang, Z.Y. | | Deposit date: | 2020-09-16 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-electron microscopy structures of VCP/p97 reveal a new mechanism of oligomerization regulation.
Iscience, 24, 2021
|
|
3LI1
 
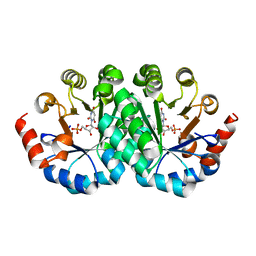 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LIX
 
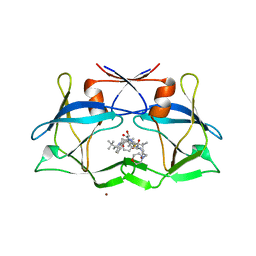 | | crystal structure of htlv protease complexed with the inhibitor KNI-10729 | | Descriptor: | N-{(1S,2S)-1-benzyl-3-[(4R)-5,5-dimethyl-4-{[(1R)-1,2,2-trimethylpropyl]carbamoyl}-1,3-thiazolidin-3-yl]-2-hydroxy-3-oxopropyl}-3-methyl-N~2~-{(2S)-2-[(morpholin-4-ylacetyl)amino]-2-phenylacetyl}-L-valinamide, Protease, ZINC ION | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
7JPN
 
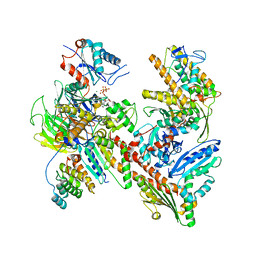 | | Cryo-EM structure of Arpin-bound Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | van Eeuwen, T, Fregoso, F.E, Dominguez, R, Zimmet, A, Boczkowska, M, Rebowski, G. | | Deposit date: | 2020-08-09 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular mechanism of Arp2/3 complex inhibition by Arpin.
Nat Commun, 13, 2022
|
|
2GEJ
 
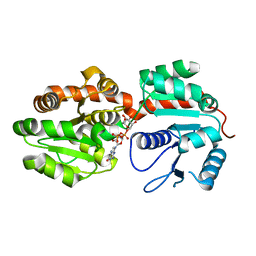 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
8P65
 
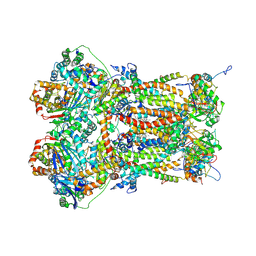 | | Cytochrome bc1 complex (Bos taurus) | | Descriptor: | Cytochrome b, Cytochrome b-c1 complex subunit 1, mitochondrial, ... | | Authors: | Phillips, B.P, Barra, I.M.C.C, Meier, T.K, Rimle, L, von Ballmoos, C. | | Deposit date: | 2023-05-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cytochrome bc1 complex (Bos taurus)
To Be Published
|
|
2GH2
 
 | |
8PSV
 
 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
7TAD
 
 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7KL9
 
 | |
7T4Q
 
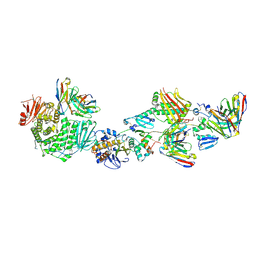 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4S
 
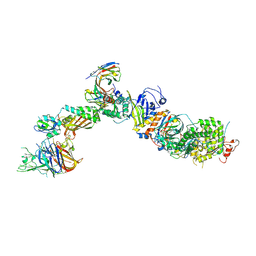 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | Authors: | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
1PHK
 
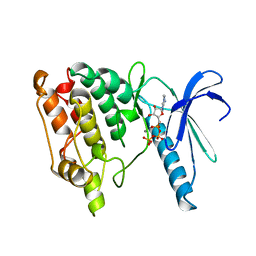 | | TWO STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHORYLASE, KINASE: AN ACTIVE PROTEIN KINASE COMPLEXED WITH NUCLEOTIDE, SUBSTRATE-ANALOGUE AND PRODUCT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE | | Authors: | Owen, D.J, Noble, M.E.M, Garman, E.F, Papageorgiou, A.C, Johnson, L.N. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Structure, 3, 1995
|
|
2MMU
 
 | | Structure of CrgA, a Cell Division Structural and Regulatory Protein from Mycobacterium tuberculosis, in Lipid Bilayers | | Descriptor: | Cell division protein CrgA | | Authors: | Das, N, Dai, J, Hung, I, Rajagopalan, M, Zhou, H, Cross, T.A. | | Deposit date: | 2014-03-18 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of CrgA, a cell division structural and regulatory protein from Mycobacterium tuberculosis, in lipid bilayers.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7SPA
 
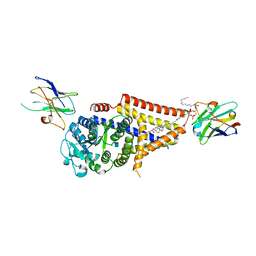 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed, channel-open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP9
 
 | | Chlorella virus Hyaluronan Synthase in the GlcNAc-primed channel-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP6
 
 | | Chlorella virus hyaluronan synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7KJX
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJV
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7SP8
 
 | | Chlorella virus Hyaluronan Synthase bound to UDP-GlcNAc | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7SP7
 
 | | Chlorella virus hyaluronan synthase inhibited by UDP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CHOLESTEROL HEMISUCCINATE, Hyaluronan synthase, ... | | Authors: | Maloney, F.P, Kuklewicz, J, Zimmer, J. | | Deposit date: | 2021-11-02 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure, substrate recognition and initiation of hyaluronan synthase.
Nature, 604, 2022
|
|
7T01
 
 | | SARS-CoV-2 S-RBD + Fab 54042-4 | | Descriptor: | 54042-4 Fab - Heavy Chain, 54042-4 Fab - Light Chain, Spike protein S1 | | Authors: | Johnson, N.V, Mclellan, J.S. | | Deposit date: | 2021-11-29 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Potent neutralization of SARS-CoV-2 variants of concern by an antibody with an uncommon genetic signature and structural mode of spike recognition.
Cell Rep, 37, 2021
|
|
