4V10
 
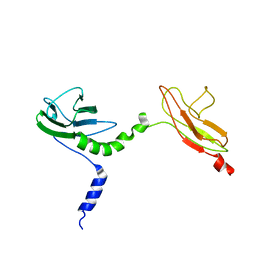 | | Skelemin Association with alpha2b,beta3 Integrin: A Structural Model | | Descriptor: | MYOMESIN-1 | | Authors: | Gorbatyuk, V, Deshmukh, L, Nguyen, K, Vinogradova, O. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Skelemin Association with Alphaiibbeta3 Integrin: A Structural Model.
Biochemistry, 53, 2014
|
|
4V2A
 
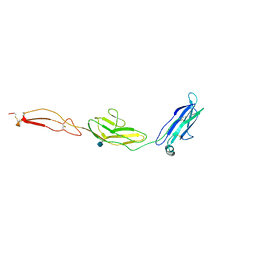 | | human Unc5A ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NETRIN RECEPTOR UNC5A | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4WV1
 
 | |
4X83
 
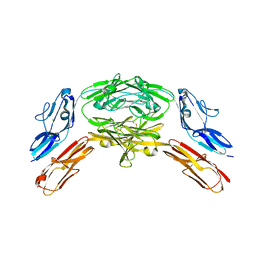 | | Crystal structure of Dscam1 isoform 7.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-10 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X8X
 
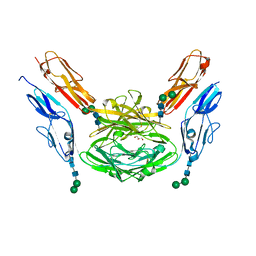 | | Crystal structure of Dscam1 isoform 1.9, N-terminal four Ig domains | | Descriptor: | Down Syndrome cell adhesion molecule isoform 1.9, GLYCEROL, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Q. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9B
 
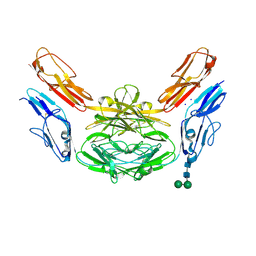 | | Crystal structure of Dscam1 isoform 4.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9F
 
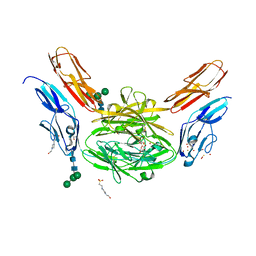 | | Crystal structure of Dscam1 isoform 6.9, N-terminal four Ig domains | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Down Syndrome Cell Adhesion Molecule isoform 6.9, GLYCEROL, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9G
 
 | | Crystal structure of Dscam1 isoform 6.44, N-terminal four Ig domains | | Descriptor: | Down Syndrome Cell Adhesion Molecule isoform 6.44, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9H
 
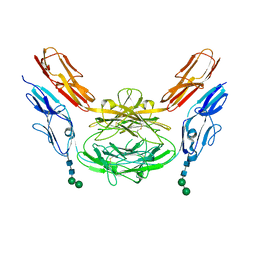 | | Crystal structure of Dscam1 isoform 8.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform AP, ... | | Authors: | Chen, Q. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4X9I
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, isoform 9.44, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-11 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB7
 
 | | Crystal structure of Dscam1 isoform 4.4, N-terminal four Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down syndrome cell adhesion molecule, isoform 4.4, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, Cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XB8
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains (with zinc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
4XHQ
 
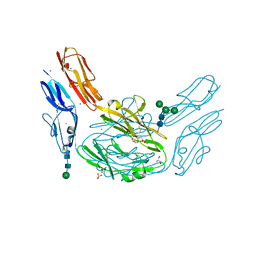 | |
4Y61
 
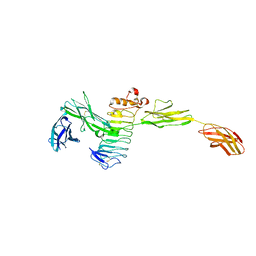 | | Crystal structure of the complex between Slitrk2 LRR1 and PTP delta Ig1-Fn1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta, SLIT and NTRK-like protein 2 | | Authors: | Yamgata, A, Sato, Y, Goto-Ito, S, Uemura, T, Maeda, A, Shiroshima, T, Yoshida, T, Fukai, S. | | Deposit date: | 2015-02-12 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.358 Å) | | Cite: | Structure of Slitrk2-PTP delta complex reveals mechanisms for splicing-dependent trans-synaptic adhesion.
Sci Rep, 5, 2015
|
|
4YFD
 
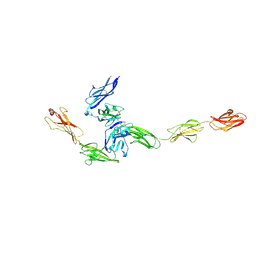 | | Crystal structure PTP delta Ig1-Fn2 in complex with IL-1RAcP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein, Receptor-type tyrosine-protein phosphatase delta | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
4YFG
 
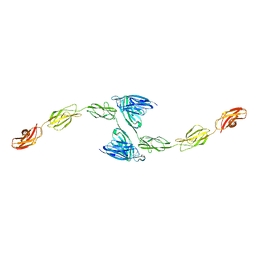 | | Crystal structure of PTP delta meA3/meB minus variant Ig1-Fn1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor-type tyrosine-protein phosphatase delta | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.491 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
4YH7
 
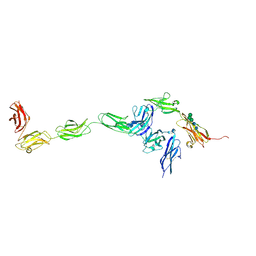 | | Crystal structure of PTPdelta ectodomain in complex with IL1RAPL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Yamagata, A, Fukai, S. | | Deposit date: | 2015-02-27 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
5AEA
 
 | |
5E4I
 
 | | Crystal structure of mouse CNTN5 Ig1-Ig4 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-5 | | Authors: | Nikolaienko, R.M, Bouyain, S. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues.
J.Biol.Chem., 291, 2016
|
|
5EO9
 
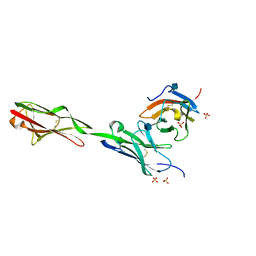 | | Crystal Structure of the complex of Dpr6 Domain 1 bound to DIP-alpha Domain 1+2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CG32791, isoform A, ... | | Authors: | Ozkan, E, Zinn, K, Garcia, K.C. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2988 Å) | | Cite: | Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins.
Cell, 163, 2015
|
|
5FM5
 
 | |
5FTT
 
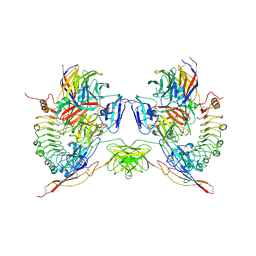 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
5I99
 
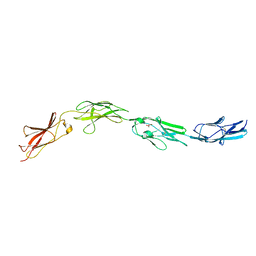 | | Crystal structure of mouse CNTN3 Ig5-Fn2 domains | | Descriptor: | Contactin-3, GLYCEROL | | Authors: | Nikolaienko, R.M, Bouyain, S. | | Deposit date: | 2016-02-19 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues.
J.Biol.Chem., 291, 2016
|
|
5JDD
 
 | |
5JDE
 
 | |
