3KLR
 
 | | Bovine H-protein at 0.88 angstrom resolution | | Descriptor: | GLYCEROL, Glycine cleavage system H protein, SULFATE ION | | Authors: | Higashiura, A, Kurakane, T, Matsuda, M, Suzuki, M, Inaka, K, Sato, M, Tanaka, H, Fujiwara, K, Nakagawa, A. | | Deposit date: | 2009-11-09 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | High-resolution X-ray crystal structure of bovine H-protein at 0.88 A resolution
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5W3F
 
 | | Yeast tubulin polymerized with GTP in vitro | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
2LP8
 
 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
4O2C
 
 | | An Nt-acetylated peptide complexed with HLA-B*3901 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-39 alpha chain, ... | | Authors: | Sun, M, Liu, J, Qi, J, Tefsen, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | N alpha-terminal acetylation for T cell recognition: molecular basis of MHC class I-restricted n alpha-acetylpeptide presentation
J.Immunol., 192, 2014
|
|
2FZ1
 
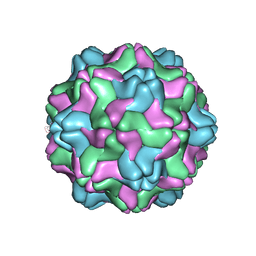 | |
3KPC
 
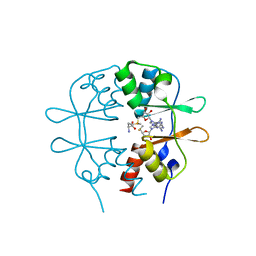 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
3KPU
 
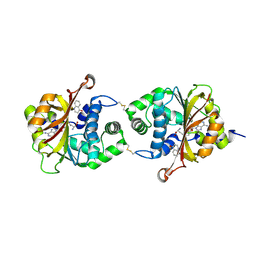 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-quinolinol | | Descriptor: | Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, quinolin-4-ol | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
5VNO
 
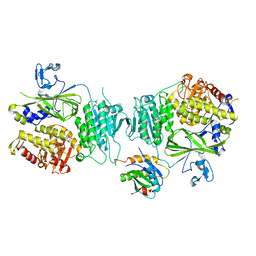 | | Crystal structure of Sec23a/Sec24a/Sec22 | | Descriptor: | Protein transport protein Sec23A, Protein transport protein Sec24A, Vesicle-trafficking protein SEC22b, ... | | Authors: | Ma, W, Goldberg, J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate.
Elife, 6, 2017
|
|
2G06
 
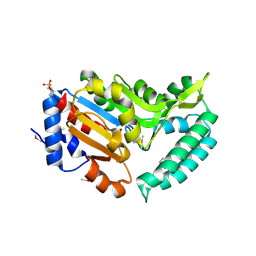 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1, with bound magnesium(II) | | Descriptor: | Cytosolic 5'-nucleotidase III, MAGNESIUM ION, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID) | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
3KR0
 
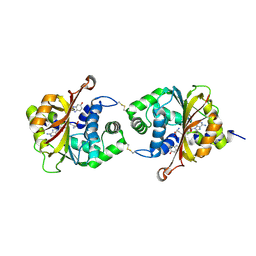 | |
3KQK
 
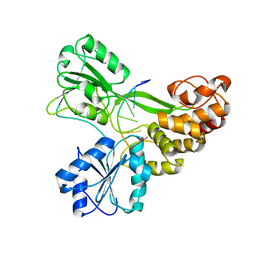 | |
4O32
 
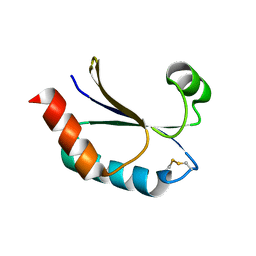 | | Structure of a malarial protein | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Egea, P.F, Koehl, A, Peng, M, Cascio, D. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystal structure and solution characterization of the thioredoxin-2 from Plasmodium falciparum, a constituent of an essential parasitic protein export complex.
Biochem.Biophys.Res.Commun., 456, 2015
|
|
3KQT
 
 | |
4O3F
 
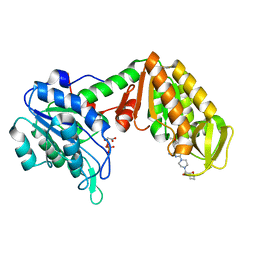 | | Crystal Structure of mouse PGK1 3PG and terazosin(TZN) ternary complex | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, Phosphoglycerate kinase 1, [4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl][(2R)-tetrahydrofuran-2-yl]methanone | | Authors: | Li, X.L, Finci, I.L, Wang, J.H. | | Deposit date: | 2013-12-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Terazosin activates Pgk1 and Hsp90 to promote stress resistance.
Nat.Chem.Biol., 11, 2015
|
|
2FQZ
 
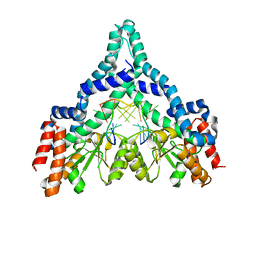 | | Metal-depleted Ecl18kI in complex with uncleaved DNA | | Descriptor: | DNA STRAND 1, DNA STRAND 2, R.Ecl18kI | | Authors: | Bochtler, M, Szczepanowski, R.H, Tamulaitis, G, Grazulis, S, Czapinska, H, Manakova, E, Siksnys, V. | | Deposit date: | 2006-01-18 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide flips determine the specificity of the Ecl18kI restriction endonuclease
Embo J., 25, 2006
|
|
7SI3
 
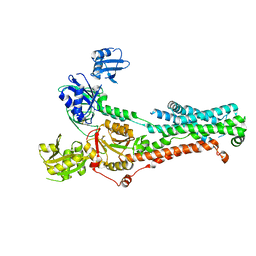 | | Consensus structure of ATP7B | | Descriptor: | MAGNESIUM ION, P-type Cu(+) transporter, TETRAFLUOROALUMINATE ION | | Authors: | Bitter, R.M, Oh, S.C, Hite, R.K, Yuan, P. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of the Wilson disease copper transporter ATP7B.
Sci Adv, 8, 2022
|
|
4NUV
 
 | |
3KTW
 
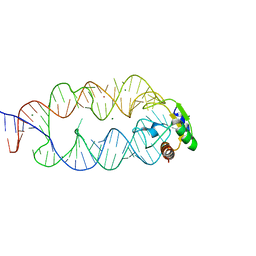 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | Authors: | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | Deposit date: | 2009-11-26 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | Descriptor: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3KW9
 
 | |
7SB2
 
 | |
4NXK
 
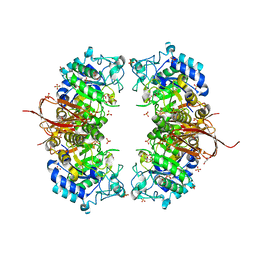 | | Crystal structure of Abp-D197A, a catalytic mutant of a GH27-b-L-arabinopyranosidase from Geobacillus stearothermophilus | | Descriptor: | Abp, a GH27 beta-L-arabinopyranosidase, CITRIC ACID, ... | | Authors: | Lansky, S, Solomon, H.V, Salama, R, Belrhali, H, Shoham, Y, Shoham, G. | | Deposit date: | 2013-12-09 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-specificity relationships in Abp, a GH27 beta-L-arabinopyranosidase from Geobacillus stearothermophilus T6
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7SAT
 
 | |
3KWZ
 
 | |
5VNF
 
 | |
