4M3V
 
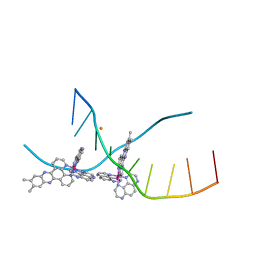 | | X-ray crystal structure of the ruthenium complex [Ru(Tap)2(dppz-{Me2})]2+ bound to d(TCGGTACCGA) | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), BARIUM ION, DNA decamer sequence | | Authors: | Niyazi, H, Teixeira, S, Mitchell, E, Forsyth, T, Cardin, C. | | Deposit date: | 2013-08-06 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of the ruthenium complex [Ru(Tap)2(dppz-{Me2})]2+ bound to d(TCGGTACCGA)
To be Published
|
|
7TRL
 
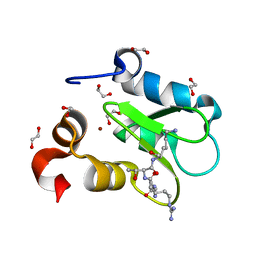 | | Crystal structure of human BIRC2 BIR3 domain in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, Histone H3, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TRM
 
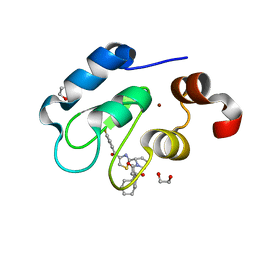 | | Crystal structure of human BIRC2 BIR3 domain in complex with inhibitor LCL-161 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, LCL-161, ... | | Authors: | Tencer, A.H, Klein, B.J, Kutateladze, T.G. | | Deposit date: | 2022-01-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for nuclear accumulation and targeting of the inhibitor of apoptosis BIRC2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1KGO
 
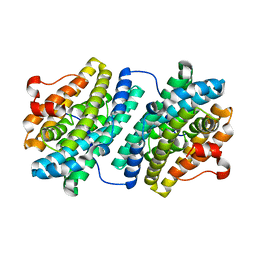 | | R2F from Corynebacterium Ammoniagenes in its reduced, Fe containing, form | | Descriptor: | FE (II) ION, Ribonucleotide reductase protein R2F | | Authors: | Hogbom, M, Huque, Y, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry, 41, 2002
|
|
8W1D
 
 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1E
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, SULFATE ION | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8W1F
 
 | | Crystal Structure of DPS-like protein PA4880 from Pseudomonas aeruginosa (dodecamer, Mg bound) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION, MAGNESIUM ION, ... | | Authors: | Lovell, S, Liu, L, Seibold, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
3ZR0
 
 | | Crystal structure of human MTH1 in complex with 8-oxo-dGMP | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION | | Authors: | Svensson, L.M, Jemth, A, Desroses, M, Loseva, O, Helleday, T, Hogbom, M, Stenmark, P. | | Deposit date: | 2011-06-13 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Mth1 and the 8-Oxo-Dgmp Product Complex.
FEBS Lett., 585, 2011
|
|
8IPF
 
 | |
3HST
 
 | |
8DTN
 
 | | The complex of nanobody 6101 with BCL11A ZF6 | | Descriptor: | B-cell lymphoma/leukemia 11A, MAGNESIUM ION, Nanobody 6101, ... | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DTU
 
 | | The complex of nanobody 5344N74D with BCL11A ZF6. | | Descriptor: | B-cell lymphoma/leukemia 11A, Nanobody 5344N74D, ZINC ION | | Authors: | Yin, M, Tenglin, K, Zhai, L, Dassama, L.M, Orkin, S.H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-02-01 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | Evolution of nanobodies specific for BCL11A.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7L9I
 
 | | Crystal structure of human ARH3-D314A bound to magnesium and ADP-ribose | | Descriptor: | ADP-ribose glycohydrolase ARH3, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Kurinov, I, Moss, J, Kim, I.K. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical analysis of human ADP-ribosyl-acceptor hydrolase 3 reveals the basis of metal selectivity and different roles for the two magnesium ions.
J.Biol.Chem., 296, 2021
|
|
7L9F
 
 | | Crystal structure of human ARH3 bound to calcium and ADP-ribose | | Descriptor: | CALCIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Kurinov, I, Moss, J, Kim, I.K. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical analysis of human ADP-ribosyl-acceptor hydrolase 3 reveals the basis of metal selectivity and different roles for the two magnesium ions.
J.Biol.Chem., 296, 2021
|
|
7L9H
 
 | | Crystal structure of human ARH3-D77A bound to magnesium and ADP-ribose | | Descriptor: | ADP-ribose glycohydrolase ARH3, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Kurinov, I, Moss, J, Kim, I.K. | | Deposit date: | 2021-01-04 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and biochemical analysis of human ADP-ribosyl-acceptor hydrolase 3 reveals the basis of metal selectivity and different roles for the two magnesium ions.
J.Biol.Chem., 296, 2021
|
|
4LP0
 
 | | Crystal structure of a topoisomerase ATP inhibitor | | Descriptor: | 6'-[(ethylcarbamoyl)amino]-4'-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridine-5-carboxylic acid, Topoisomerase IV subunit B | | Authors: | Basarab, G.S, Manchester, J.I, Bist, S, Boriack-Sjodin, P.A, Dangel, B, Illingsworth, R, Uria-Nickelsen, M, Sherer, B.A, Sriram, S, Eakin, A.E. | | Deposit date: | 2013-07-14 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
4LPB
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | Descriptor: | 1-ethyl-3-{5'-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)-4-[4-(trifluoromethyl)-1,3-thiazol-2-yl]-3,3'-bipyridin-6-yl}urea, Topoisomerase IV subunit B | | Authors: | Boriack-Sjodin, A. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-to-Hit-to-Lead Discovery of a Novel Pyridylurea Scaffold of ATP Competitive Dual Targeting Type II Topoisomerase Inhibiting Antibacterial Agents.
J.Med.Chem., 56, 2013
|
|
5A7L
 
 | |
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | Descriptor: | IODIDE ION, Protein Ten1 | | Authors: | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | Deposit date: | 2009-09-25 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3UVU
 
 | |
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
4QAG
 
 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
7MSV
 
 | |
7MPZ
 
 | |
1MH9
 
 | | Crystal Structure Analysis of deoxyribonucleotidase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, deoxyribonucleotidase | | Authors: | Rinaldo-Matthis, A, Rampazzo, C, Reichard, P, Bianchi, V, Nordlund, P. | | Deposit date: | 2002-08-19 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a human mitochondrial deoxyribonucleotidase.
Nat.Struct.Biol., 9, 2002
|
|
