7ZKW
 
 | |
7SZQ
 
 | | Human P300 complexed with an azaindazole inhibitor | | Descriptor: | 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-1H-pyrazolo[4,3-b]pyridin-5-yl-D-prolinamide, Histone acetyltransferase p300 | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-29 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SSK
 
 | | Human P300 complexed with a glycine-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SS8
 
 | | Human P300 complexed with a proline-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(4-chlorophenyl)cyclopentane-1-carbonyl]-N-{[3-(methylcarbamoyl)phenyl]methyl}-D-prolinamide, Histone acetyltransferase p300, ... | | Authors: | Shewchuk, L.M, Reid, R.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Proline-Based p300/CBP Inhibitors Using DNA-Encoded Library Technology in Combination with High-Throughput Screening.
J.Med.Chem., 65, 2022
|
|
7SQ5
 
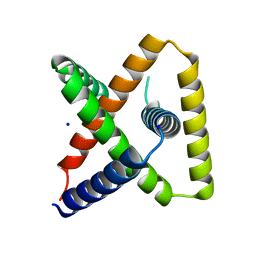 | | Designed trefoil knot protein, variant 3 | | Descriptor: | Designed trefoil knot protein, variant 3, SODIUM ION | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SUR
 
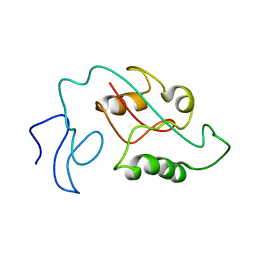 | |
7T6D
 
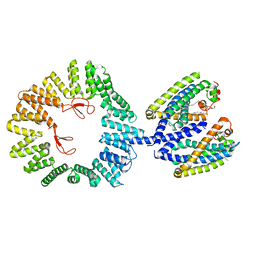 | | CryoEM structure of the YejM/LapB complex | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Inner membrane protein YejM, ... | | Authors: | Mi, W, Shu, S. | | Deposit date: | 2021-12-13 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Regulatory mechanisms of lipopolysaccharide synthesis in Escherichia coli.
Nat Commun, 13, 2022
|
|
7ZOF
 
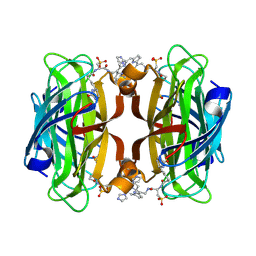 | | Streptavidin Iron-Porphyrin | | Descriptor: | (2R)-2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-3-[[(5Z,10Z,14Z,19Z)-15-[[[(2R)-2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]-3-sulfo-propanoyl]amino]methyl]-1,4,21,23-tetrahydroporphyrin-5-yl]methylamino]-3-oxidanylidene-propane-1-sulfonic acid, FE (III) ION, IMIDAZOLE, ... | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2022-04-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Streptavidin Iron-Porphyrin
To Be Published
|
|
7ZJT
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis at 1.96 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, CHLORIDE ION | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7ZYF
 
 | | Insulin regulated aminopeptidase (IRAP) in complex with a nanomolar alpha hydroxy beta amino acid based inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mpakali, A, Stratikos, E, Giastas, P, Papakyriakou, A. | | Deposit date: | 2022-05-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
7ZM2
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclophostin-like inhibitor CyC8b | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, methoxy-[(3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]pentadecyl]phosphinic acid | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
7SUQ
 
 | |
7ZM3
 
 | | Crystal structure of HsaD from Mycobacterium tuberculosis in complex with Cyclipostin-like inhibitor CyC17 | | Descriptor: | 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, SULFATE ION, hexadecyl dihydrogen phosphate | | Authors: | Barelier, S, Roig-Zamboni, V, Cavalier, J.F, Sulzenbacher, G. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Direct capture, inhibition and crystal structure of HsaD (Rv3569c) from M. tuberculosis.
Febs J., 290, 2023
|
|
8A5Q
 
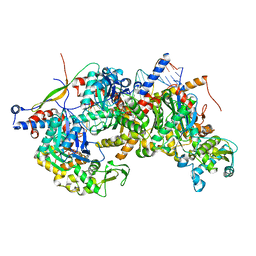 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5P
 
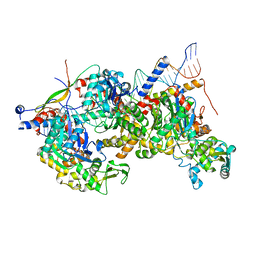 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on curved DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7TC3
 
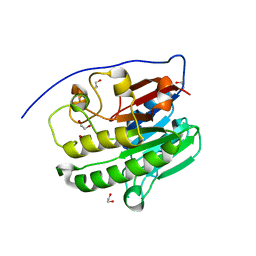 | | Human APE1 in the apo form | | Descriptor: | 1,2-ETHANEDIOL, DNA-(apurinic or apyrimidinic site) endonuclease, mitochondrial | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
7ZX8
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (OC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TC2
 
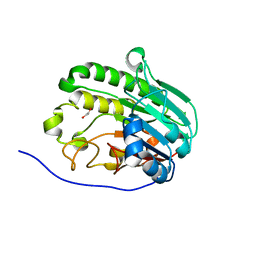 | | Human APE1 in complex with 5-nitroindole-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 5-nitro-1H-indole-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2021-12-22 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Characterizing inhibitors of human AP endonuclease 1.
Plos One, 18, 2023
|
|
8A5O
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80 | | Descriptor: | Actin, Actin-like protein ARP8, Actin-related protein 4, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7TCV
 
 | | VDAC K12E mutant | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Voltage-dependent anion-selective channel protein 1 | | Authors: | Khan, F, Abramson, J. | | Deposit date: | 2021-12-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Dynamical control of the mitochondrial beta-barrel channel VDAC by electrostatic and mechanical coupling
To Be Published
|
|
8A5D
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | Descriptor: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
7TAY
 
 | | Bos Taurus Mitochondrial BC1 in complex with Pyramoxadone | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-3-anilino-5-methyl-5-(6-phenoxypyridin-3-yl)-1,3-oxazolidine-2,4-dione, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Zhou, F, Esser, L. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conformation Switch of Rieske ISP subunit is revealed by the Crystal Structure of Bacterial Cytochrome bc1 in Complex with Azoxystrobin
to be published
|
|
7ZTD
 
 | | Non-muscle F-actin decorated with non-muscle tropomyosin 3.2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Non-muscle tropomyosin 3.2, actin, ... | | Authors: | Selvaraj, M, Kokate, S, Kogan, K, Kotila, T, Kremneva, E, Lappalainen, P, Huiskonen, J.T. | | Deposit date: | 2022-05-09 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis underlying specific biochemical activities of non-muscle tropomyosin isoforms.
Cell Rep, 42, 2023
|
|
7TCE
 
 | | Crystal structure of delta sub IV Rhodobacter Sphaeroides bc1 with the antimalarial drug atovaquone. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxynaphthalene-1,4-dione, Cytochrome b, ... | | Authors: | Esser, L, Xia, D, Zhou, F. | | Deposit date: | 2021-12-23 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of delta sub IV Rhodobacter Sphaeroides bc1 with the antimalarial drug atovaquone.
To Be Published
|
|
