7RV0
 
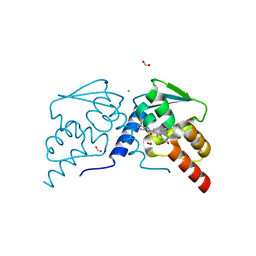 | |
7RV2
 
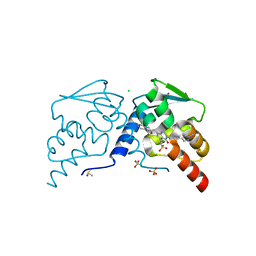 | |
7RUW
 
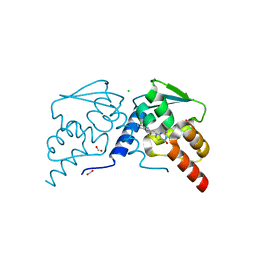 | | Crystal structure of the BCL6 BTB domain in complex with OICR-7859 | | Descriptor: | 2-(2-amino-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-N-(3-chloropyridin-4-yl)acetamide, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-7859
To Be Published
|
|
7RUZ
 
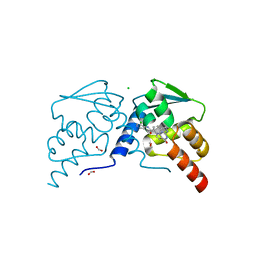 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8445 | | Descriptor: | 2-[2-(benzylamino)-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-(3-chloropyridin-4-yl)acetamide, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8445
To Be Published
|
|
7RV1
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8826 | | Descriptor: | (E)-3-(7-(2-((3-chloropyridin-4-yl)amino)-2-oxoethyl)-3-(3-(1-methyl-1H-pyrazol-4-yl)prop-2-yn-1-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)acrylamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8826
To Be Published
|
|
7RUY
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8388 | | Descriptor: | B-cell lymphoma 6 protein, FORMIC ACID, N-(3-chloropyridin-4-yl)-2-(2-{[(2,4-dimethoxyphenyl)methyl]amino}-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8388
To Be Published
|
|
4V6X
 
 | | Structure of the human 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
1UMU
 
 | |
8Y0W
 
 | | dormant ribosome with eIF5A, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
8Y0X
 
 | | Dormant ribosome with SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Du, M, Zeng, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Implication of Stm1 in the protection of eIF5A, eEF2 and tRNA through dormant ribosomes.
Front Mol Biosci, 11, 2024
|
|
4UY7
 
 | |
4UY6
 
 | |
6LUR
 
 | |
4V55
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin and ribosome recycling factor (RRF). | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
3KUP
 
 | | Crystal Structure of the CBX3 Chromo Shadow Domain | | Descriptor: | Chromobox protein homolog 3, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Li, Z, Li, Y, Kozieradzki, I, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-27 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the CBX3 Chromo Shadow Domain
to be published
|
|
4WOI
 
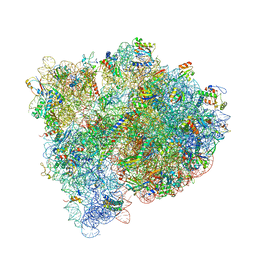 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
4V5B
 
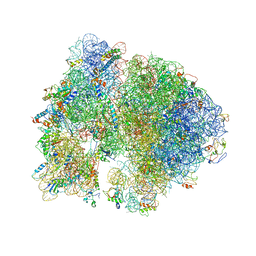 | | Structure of PDF binding helix in complex with the ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Bingel-Erlenmeyer, R, Kohler, R, Kramer, G, Sandikci, A, Antolic, S, Maier, T, Schaffitzel, C, Wiedmann, B, Bukau, B, Ban, N. | | Deposit date: | 2007-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | A Peptide Deformylase-Ribosome Complex Reveals Mechanism of Nascent Chain Processing.
Nature, 452, 2008
|
|
4V7K
 
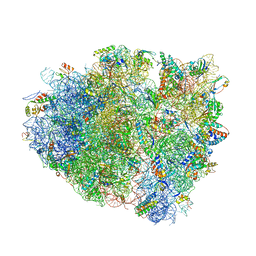 | | Structure of RelE nuclease bound to the 70S ribosome (postcleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
8T5H
 
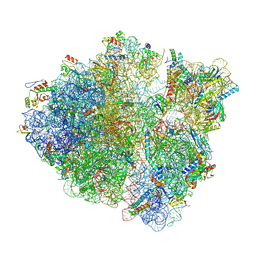 | | Cryo-EM studies of the interplay between uS2 ribosomal protein and leaderless mRNA during bacterial translation initiation | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Gottesman, M.E, Frank, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | How Dedicated Ribosomes Translate a Leaderless mRNA.
J.Mol.Biol., 436, 2024
|
|
8T5D
 
 | | Cryo-EM studies of the interplay between uS2 ribosomal protein and leaderless mRNA during bacterial translation initiation | | Descriptor: | 16s RNA, 23S RNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Gottesman, M.E, Frank, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | How Dedicated Ribosomes Translate a Leaderless mRNA.
J.Mol.Biol., 436, 2024
|
|
8R3V
 
 | |
8BPA
 
 | | Cryo-EM structure of the human SIN3B histone deacetylase complex at 3.7 Angstrom | | Descriptor: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | Authors: | Wan, M.S.M, Muhammad, R, Koliopolous, M.G, Alfieri, C. | | Deposit date: | 2022-11-16 | | Release date: | 2023-05-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
3N5E
 
 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | Descriptor: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6XRE
 
 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
7WA4
 
 | | Crystal structure of GIGANTEA in complex with LKP2 | | Descriptor: | Adagio protein 2, FLAVIN MONONUCLEOTIDE, Protein GIGANTEA | | Authors: | Pathak, D, Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the regulation of blue-light receptors by GIGANTEA.
Cell Rep, 39, 2022
|
|
