4HSW
 
 | | Structure of the L100F mutant of dehaloperoxidase-hemoglobin A from Amphitrite ornata | | Descriptor: | Dehaloperoxidase A, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Thompson, M.K, Plummer, A, Franzen, S. | | Deposit date: | 2012-10-31 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Role of polarity of the distal pocket in the control of inhibitor binding in dehaloperoxidase-hemoglobin.
Biochemistry, 52, 2013
|
|
5PHC
 
 | | PanDDA analysis group deposition -- Crystal Structure of JMJD2D in complex with N09649a | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-hydroxyphenyl)phenol, Lysine-specific demethylase 4D, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Szykowska, A, Burgess-Brown, N, Brennan, P.E, Cox, O, Oppermann, U, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4HSJ
 
 | | 1.88 angstrom x-ray crystal structure of piconlinic-bound 3-hydroxyanthranilate-3,4-dioxygenase | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION, PYRIDINE-2-CARBOXYLIC ACID | | Authors: | Liu, F, Chen, L, Davis, C.I, Liu, A. | | Deposit date: | 2012-10-30 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | An Iron Reservoir to the Catalytic Metal: THE RUBREDOXIN IRON IN AN EXTRADIOL DIOXYGENASE.
J.Biol.Chem., 290, 2015
|
|
2RHR
 
 | | P94L actinorhodin ketordeuctase mutant, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-09 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
4HUQ
 
 | | Crystal Structure of a transporter | | Descriptor: | Energy-coupling factor transporter ATP-binding protein EcfA 1, Energy-coupling factor transporter ATP-binding protein EcfA 2, Energy-coupling factor transporter transmembrane protein EcfT, ... | | Authors: | Zhang, P, Xu, K, Zhang, M, Zhao, Q, Yu, F. | | Deposit date: | 2012-11-03 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis.
Nature, 497, 2013
|
|
4I25
 
 | | 2.00 Angstroms X-ray crystal structure of NAD- and substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-amino-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I3J
 
 | | Structures of PR1 intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected AT 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4HMX
 
 | | Crystal structure of PhzG from Burkholderia lata 383 in complex with tetrahydrophenazine-1-carboxylic acid | | Descriptor: | (1R,10aS)-1,2,10,10a-tetrahydrophenazine-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, Pyridoxamine 5'-phosphate oxidase | | Authors: | Xu, N.N, Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4I8J
 
 | | Bovine trypsin at 0.87 A resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IA9
 
 | | Crystal structure of human WD REPEAT DOMAIN 5 in complex with 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-4-fluoro-3-methyl-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Bolshan, Y, Getlik, M, Tempel, W, Kuznetsova, E, Wasney, G.A, Hajian, T, Poda, G, Nguyen, K.T, Schapira, M, Brown, P.J, Al-awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Smil, D, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-06 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction.
ACS Med Chem Lett, 4, 2013
|
|
4I2R
 
 | | 2.15 Angstroms X-ray crystal structure of NAD- and alternative substrate-bound 2-aminomuconate 6-semialdehyde dehydrogenase from Pseudomonas fluorescens | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Davis, I, Chen, L, Liu, A. | | Deposit date: | 2012-11-22 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
4I3P
 
 | |
4I41
 
 | | Crystal Structure of human Ser/Thr kinase Pim1 in complex with mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Wan, X, Xie, Y, Huang, N. | | Deposit date: | 2012-11-27 | | Release date: | 2013-12-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new target for an old drug: identifying mitoxantrone as a nanomolar inhibitor of PIM1 kinase via kinome-wide selectivity modeling.
J. Med. Chem., 56, 2013
|
|
4HSL
 
 | | 2.00 angstrom x-ray crystal structure of substrate-bound E110A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Liu, F, Chen, L, Liu, A. | | Deposit date: | 2012-10-30 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 angstrom x-ray crystal structure of substrate-bound
E110A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus
metallidurans
To be Published
|
|
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4I39
 
 | | Structures of ICT and PR1 intermediates from time-resolved laue crystallography collected at 14ID-B, APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4I06
 
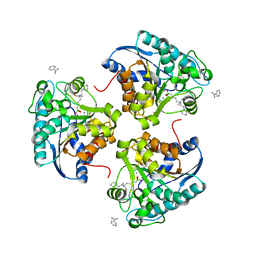 | | Crystal structure of human Arginase-2 complexed with inhibitor 14 | | Descriptor: | Arginase-2, mitochondrial, BENZAMIDINE, ... | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Whitehouse, D.L, Golebiowski, A, Ji, M, Zhang, M, Beckett, P, Sheeler, R, Andreoli, M, Conway, B, Mahboubi, K, Schroeter, H, Van Zandt, M.C, Podjarny, A. | | Deposit date: | 2012-11-16 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-2-Amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic Acid and Congeners As Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury.
J.Med.Chem., 56, 2013
|
|
4I8X
 
 | | Crystal structure of rabbit LDHA in complex with AP27460 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-04 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4IAU
 
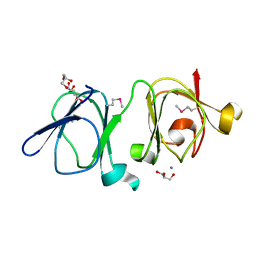 | | Atomic resolution structure of Geodin, a beta-gamma crystallin from Geodia cydonium | | Descriptor: | Beta-gamma-crystallin, CALCIUM ION, GLYCEROL | | Authors: | Vergara, A, Grassi, M, Sica, F, Mazzarella, L, Merlino, A. | | Deposit date: | 2012-12-07 | | Release date: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A novel interdomain interface in crystallins: structural characterization of the [beta][gamma]-crystallin from Geodia cydonium at 0.99 A resolution
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4IFC
 
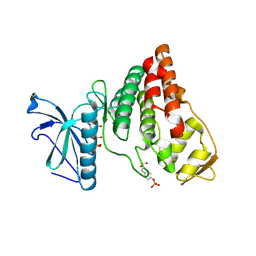 | | Crystal Structure of ADP-bound Human PRPF4B kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-14 | | Release date: | 2013-08-28 | | Last modified: | 2013-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
4IJ6
 
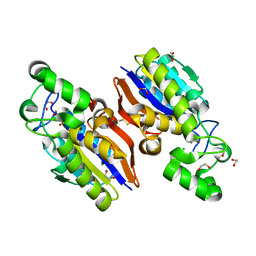 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase Mutant (H9A) from Hydrogenobacter thermophilus TK-6 in Complex with L-phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOSERINE, ... | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
4ION
 
 | | Macrolepiota procera ricin B-like lectin (MPL) | | Descriptor: | GLYCEROL, Ricin B-like lectin | | Authors: | Renko, M, Zurga, S, Sabotic, J, Pohleven, J, Kos, J, Turk, D. | | Deposit date: | 2013-01-08 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Macrolepiota procera ricin B-like lectin (MPL)
TO BE PUBLISHED
|
|
4IOH
 
 | | Crystal Structure of the Tll1086 protein from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR258 | | Descriptor: | Tll1086 protein | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Maglaqui, M, Xiao, X, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-01-07 | | Release date: | 2013-01-30 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal Structure of the Tll1086 protein from Thermosynechococcus elongatus.
To be Published
|
|
4IH9
 
 | | Crystal structure of rice DWARF14 (D14) | | Descriptor: | Dwarf 88 esterase | | Authors: | Zhou, X.E, Zhao, L.-H, Wu, Z.-S, Yi, W, Li, S, Li, Y, Xu, Y, Xu, T.-H, Liu, Y, Chen, R.-Z, Kovach, A, Kang, Y, Hou, L, He, Y, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2012-12-18 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two phytohormone signal-transducing alpha / beta hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14.
Cell Res., 23, 2013
|
|
4IJB
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR288 | | Descriptor: | Engineered Protein OR288 | | Authors: | Vorobiev, S, Su, M, Bick, M.J, Seetharaman, J, Khare, S, Maglaqui, M, Xiao, R, Lee, D, Day, A, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (1.739 Å) | | Cite: | Crystal Structure of Engineered Protein OR288.
To be Published
|
|
