7PRD
 
 | |
6YMK
 
 | | Crystal structure of the SAM-SAH riboswitch with AMP | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
7PRE
 
 | |
4XRG
 
 | |
4XS0
 
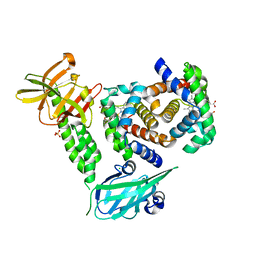 | | Human methemoglobin in complex with the second and third NEAT domains of IsdH(F365Y/A369F/Y642A) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Dickson, C.F, Jacques, D.A, Guss, J.M, Gell, D.A. | | Deposit date: | 2015-01-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of haemoglobin bound to the haemoglobin receptor IsdH from Staphylococcus aureus shows disruption of the native alpha-globin haem pocket.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7WAF
 
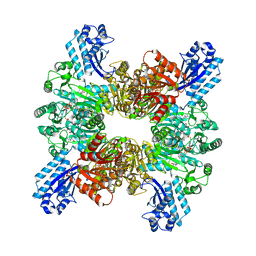 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS and 4x(beta-Asp-Arg) | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, Cyanophycin synthase, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
4XTO
 
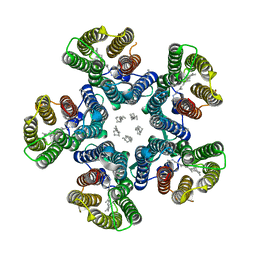 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric red form, pH 5.6 | | Descriptor: | EICOSANE, SODIUM ION, Sodium pumping rhodopsin | | Authors: | Gushchin, I, Shevchenko, V, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2015-01-23 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a light-driven sodium pump.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7WDL
 
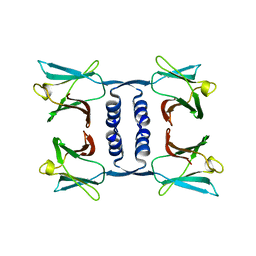 | | Fungal immunomodulatory protein FIP-nha | | Descriptor: | Fungal immunomodulatory protein | | Authors: | Liu, Y, Bastiaan-Net, S, Zhang, Y, Hoppenbrouwers, T, Xie, Y, Wang, Y, Wei, X, Du, G, Zhang, H, Imam, K.M.S.U, Wichers, H.J, Li, Z. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Linking the thermostability of FIP-nha (Nectria haematococca) to its structural properties.
Int.J.Biol.Macromol., 213, 2022
|
|
6Y6B
 
 | | Crystal structure of human 14-3-3 gamma in complex with CaMKK2 14-3-3 binding motif Ser100 and 16-OMe-Fusicoccin H | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[[(1~{S},3~{R},6~{S},7~{S},8~{R},9~{R},10~{R},11~{R},14~{S})-14-(me thoxymethyl)-3,10-dimethyl-9-oxidanyl-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradecanyl]oxy]oxane-3,4,5-triol, 14-3-3 protein gamma, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Lentini Santo, D, Obsilova, V, Obsil, T. | | Deposit date: | 2020-02-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Stabilization of Protein-Protein Interactions between CaMKK2 and 14-3-3 by Fusicoccins.
Acs Chem.Biol., 15, 2020
|
|
7SUR
 
 | |
8DBZ
 
 | |
4XYY
 
 | |
7WAC
 
 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) | | Descriptor: | Cyanophycin synthase | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7Q9S
 
 | | Crystal structure of PDE6D KRas peptide complex with Compound-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3-(1~{H}-pyrazol-5-yl)imidazo[1,2-a]pyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Yelland, T, Ismail, S. | | Deposit date: | 2021-11-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
J.Med.Chem., 65, 2022
|
|
7WAD
 
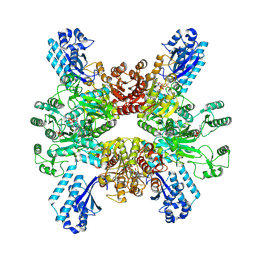 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS | | Descriptor: | Cyanophycin synthase, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Kawasaki, M, Miyakawa, T, Yang, J, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
6PEQ
 
 | |
8JB3
 
 | |
7Q9Q
 
 | |
7Q9R
 
 | | Cocrystal structure of PDE6D bound to NRAS peptide | | Descriptor: | 1,2-ETHANEDIOL, FARNESYL, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ... | | Authors: | Yelland, T, Ismail, S. | | Deposit date: | 2021-11-14 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
J.Med.Chem., 65, 2022
|
|
7SUQ
 
 | |
6PIJ
 
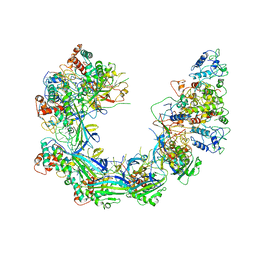 | | Target DNA-bound V. cholerae TniQ-Cascade complex, closed conformation | | Descriptor: | Non-targeting strand ssDNA, Targeting strand ssDNA, TniQ monomer 2, ... | | Authors: | Halpin-Healy, T, Klompe, S, Sternberg, S.H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Nature, 577, 2020
|
|
4Y18
 
 | |
7WAE
 
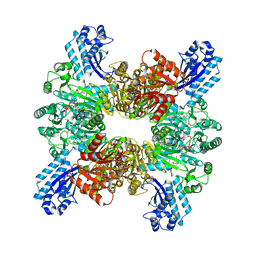 | | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) with ATPgammaS, 4x(beta-Asp-Arg), and aspartate | | Descriptor: | 4x(beta-Asp-Arg), ARGININE, ASPARTIC ACID, ... | | Authors: | Miyakawa, T, Yang, J, Kawasaki, M, Adachi, N, Fujii, A, Miyauchi, Y, Muramatsu, T, Moriya, T, Senda, T, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-09-07 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase.
Nat Commun, 13, 2022
|
|
7QF9
 
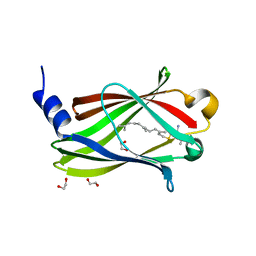 | | Crystal structure of PDE6D bound to HRas peptide | | Descriptor: | 1,2-ETHANEDIOL, FARNESYL, HRas peptide, ... | | Authors: | Yelland, T, Ismail, I. | | Deposit date: | 2021-12-05 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
J.Med.Chem., 65, 2022
|
|
8CWR
 
 | |
