6HTZ
 
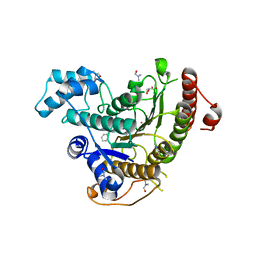 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 8 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-(2-phenylethanoylamino)benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6B6U
 
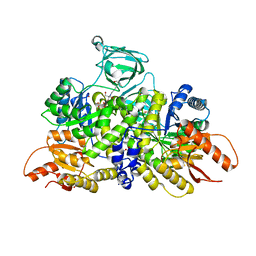 | | Pyruvate Kinase M2 mutant - S437Y | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Srivastava, D, Dey, M. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of a Dimeric Variant of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 56, 2017
|
|
6B85
 
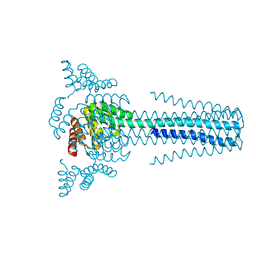 | | Crystal structure of transmembrane protein TMHC4_R | | Descriptor: | TMHC4_R | | Authors: | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.889 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B4W
 
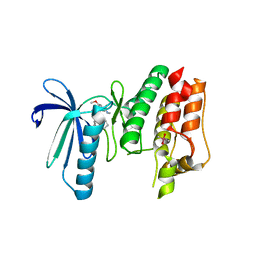 | | TTK in Complex with Inhibitor | | Descriptor: | 4-{[4-(cyclopentyloxy)-5-(2-methyl-1,3-benzoxazol-6-yl)-7H-pyrrolo[2,3-d]pyrimidin-2-yl]amino}-3-methoxy-N-methylbenzamide, CACODYLATE ION, Dual specificity protein kinase TTK | | Authors: | Delker, S, Chamberlain, P.P. | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Discovery of a Dual TTK Protein Kinase/CDC2-Like Kinase (CLK2) Inhibitor for the Treatment of Triple Negative Breast Cancer Initiated from a Phenotypic Screen.
J. Med. Chem., 60, 2017
|
|
3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
4HNN
 
 | |
6BEW
 
 | | Solution structure of de novo macrocycle design7.2 | | Descriptor: | (DHI)P(DAS)(DGN)(DSN)(DGL)P | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | Descriptor: | TMHC2_E | | Authors: | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | Deposit date: | 2017-10-05 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.947 Å) | | Cite: | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6BET
 
 | | Solution structure of de novo macrocycle design12_ss | | Descriptor: | H(DPR)(DVA)CIP(DPR)E(DLY)VC(DGL) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6JO6
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
4QEL
 
 | | Crystal Structure of Benzoylformate Decarboxylase Mutant H70A | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Andrews, F.H, Rogers, M.P, Brodkin, H.R, McLeish, M.J. | | Deposit date: | 2014-05-16 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Structural investigation of benzoylformate decarboxylase active site variants
To be Published
|
|
6BF5
 
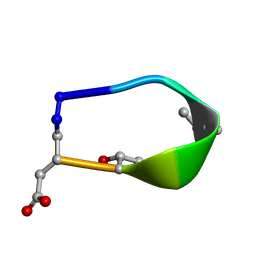 | | Solution structure of de novo macrocycle design7.3a | | Descriptor: | QDP(DPR)K(DTH)(DAS) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEO
 
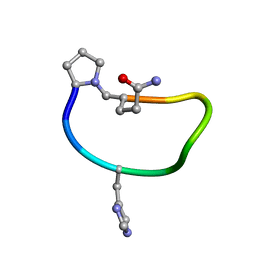 | | Solution structure of de novo macrocycle design9.1 | | Descriptor: | (DPR)PY(DHI)PKDL(DGN) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BER
 
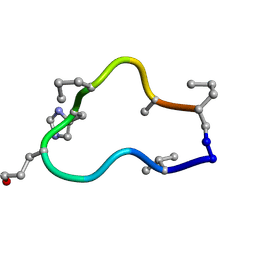 | | Solution structure of de novo macrocycle design10.2 | | Descriptor: | E(DVA)DP(DGL)(DHI)(DPR)N(DAL)(DPR) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEQ
 
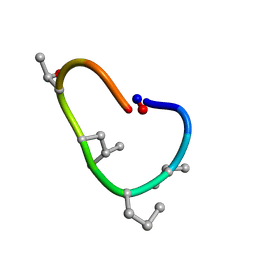 | | Solution structure of de novo macrocycle design10.1 | | Descriptor: | AAR(DVA)(DPR)R(DLE)(DTH)PE | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEU
 
 | | Solution structure of de novo macrocycle design14_ss | | Descriptor: | (DCY)N(DVA)(DPR)DVYC(DPR)(DSG)KY(DVA)(DPR) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | Deposit date: | 2017-10-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BE9
 
 | | Solution structure of de novo macrocycle design7.1 | | Descriptor: | T(DLY)NDT(DSG)(DPR) | | Authors: | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, B. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
3VMW
 
 | | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate | | Descriptor: | Pectate lyase, SULFATE ION, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Zheng, Y, Huang, C.H, Liu, W, Ko, T.P, Xue, Y, Zhou, C, Zhang, G, Guo, R.T, Ma, Y. | | Deposit date: | 2011-12-17 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and substrate-binding mode of a novel pectate lyase from alkaliphilic Bacillus sp. N16-5.
Biochem.Biophys.Res.Commun., 420, 2012
|
|
6HSK
 
 | | Crystal structure of a human HDAC8 L6 loop mutant complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Ramos-Morales, E, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSZ
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 2 | | Descriptor: | 3-benzamido-4-methyl-~{N}-oxidanyl-benzamide, GLYCEROL, Histone deacetylase, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
4MB7
 
 | |
6HVG
 
 | |
4P6F
 
 | |
4CVX
 
 | | COMPLEX OF A B2 CHICKEN MHC CLASS I MOLECULE AND A 9MER CHICKEN PEPTIDE | | Descriptor: | BETA-2-MICROGLOBULIN, MHC CLASS I ALPHA CHAIN 2, SELF-PEPTIDE | | Authors: | Chappell, P.E, Roversi, P, Harrison, M.C, Mears, L.E, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2014-03-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Elife, 4, 2015
|
|
2HQP
 
 | |
