6H7F
 
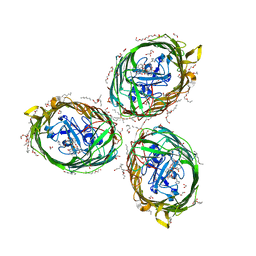 | | Crystal structure of BauA, the Ferric preacinetobactin receptor from Acinetobacter baumannii in complex with Fe3+-Preacinetobactin-acinetobactin | | Descriptor: | (4~{S},5~{R})-2-[2,3-bis(oxidanyl)phenyl]-~{N}-[2-(1~{H}-imidazol-4-yl)ethyl]-5-methyl-~{N}-oxidanyl-4,5-dihydro-1,3-oxazole-4-carboxamide, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,2-ETHANEDIOL, ... | | Authors: | Moynie, L, Naismith, J.H. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogenAcinetobacter baumannii.
Elife, 7, 2018
|
|
6H7A
 
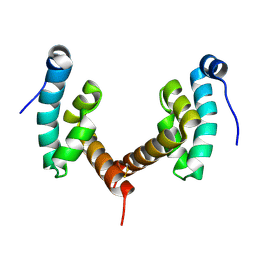 | |
6APA
 
 | |
6H54
 
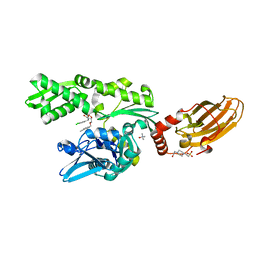 | | CRYSTAL STRUCTURE OF BOVINE HSC70(AA1-554)E213A/D214A IN COMPLEX WITH INHIBITOR VER155008 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, GLYCEROL, ... | | Authors: | Plank, C, Zehe, M, Grimm, C, Sotriffer, C. | | Deposit date: | 2018-07-23 | | Release date: | 2019-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Combined In-Solution Fragment Screening and Crystallographic Binding-Mode Analysis with a Two-Domain Hsp70 Construct.
Acs Chem.Biol., 2024
|
|
6GF2
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
6GFT
 
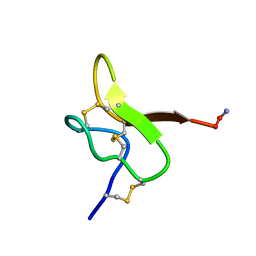 | |
6H8O
 
 | |
6H8S
 
 | |
6H7V
 
 | |
6H90
 
 | |
6HB9
 
 | |
6DCN
 
 | | Bcl-xL complex with Beclin 1 BH3 domain T108pThr | | Descriptor: | BCL-xl protein, Beclin-1 | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DGK
 
 | |
6DAD
 
 | | 1.65 Angstrom crystal structure of the N97I Ca/CaM:CaV1.2 IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Voltage-dependent L-type calcium channel subunit alpha-1C | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DF9
 
 | | Kaiso (ZBTB33) E535Q zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6DJW
 
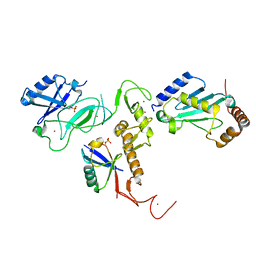 | | Crystal Structure of pParkin (REP and RING2 deleted)-pUb-UbcH7 complex | | Descriptor: | RBR-type E3 ubiquitin transferase,RBR-type E3 ubiquitin transferase, Ubiquitin, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Sauve, V, Sung, G, Trempe, J.F, Gehring, K. | | Deposit date: | 2018-05-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Mechanism of parkin activation by phosphorylation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DL0
 
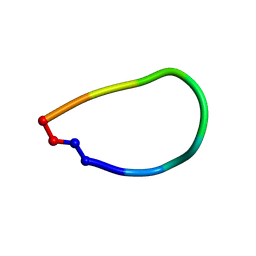 | | Crystal structure of pohlianin C, an orbitide from Jatropha pohliana | | Descriptor: | pohlianin C | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6DCO
 
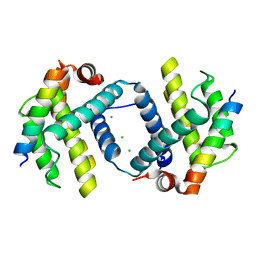 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DN6
 
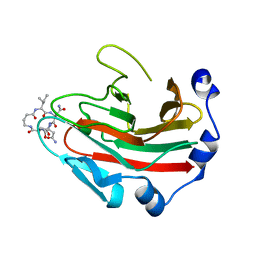 | |
6DHA
 
 | |
6DGL
 
 | |
6DKZ
 
 | | Racemic structure of ribifolin, an orbitide from Jatropha ribifolia | | Descriptor: | ribifolin | | Authors: | Wang, C.K, King, G.J, Ramalho, S.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-11-14 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Synthesis, Racemic X-ray Crystallographic, and Permeability Studies of Bioactive Orbitides from Jatropha Species.
J. Nat. Prod., 81, 2018
|
|
6DNS
 
 | | Endo-fucoidan hydrolase MfFcnA9 from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
6DSL
 
 | | Consensus engineered intein (Cat) with atypical split site | | Descriptor: | Consensus engineered intein CatC, Consensus engineered intein CatN | | Authors: | Sekar, G, Stevens, A.J, Muir, T.W, Cowburn, D. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An Atypical Mechanism of Split Intein Molecular Recognition and Folding.
J. Am. Chem. Soc., 140, 2018
|
|
6DAH
 
 | | 2.5 Angstrom crystal structure of the N97S CaM mutant | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Wang, K, Van Petegem, F. | | Deposit date: | 2018-05-01 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
