6GXY
 
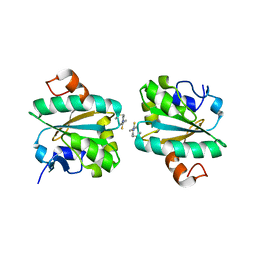 | | Tryparedoxin from Trypanosoma brucei in complex with CFT | | Descriptor: | 5-(4-fluorophenyl)-2-methyl-3~{H}-thieno[2,3-d]pyrimidin-4-one, Tryparedoxin | | Authors: | Bader, N, Wagner, A, Hellmich, U, Schindelin, H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6GY9
 
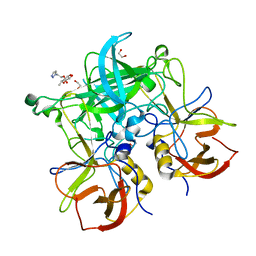 | | Fucose-functionalized precision glycomacromolecules targeting human norovirus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,3-triazol-1-yl)ethyl 6-deoxy-alpha-L-galactopyranoside, Capsid protein | | Authors: | Ruoff, K, Kilic, T, Hansman, G.S. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fucose-Functionalized Precision Glycomacromolecules Targeting Human Norovirus Capsid Protein.
Biomacromolecules, 19, 2018
|
|
6D12
 
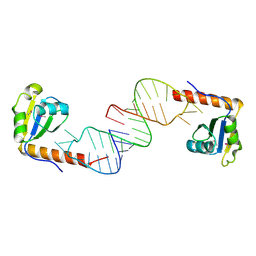 | | Crystal structure of C-terminal xRRM domain of human Larp7 bound to 7SK stem-loop 4 RNA | | Descriptor: | La-related protein 7, human 7SK RNA stem-loop 4 | | Authors: | Eichhorn, C.D, Cascio, D, Sawaya, M.R, Feigon, J. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CDH
 
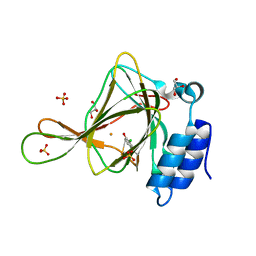 | | Crystal structure of ferrous form of the Cl-Tyr157 human cysteine dioxygenase with both uncrosslinked and crosslinked cofactor | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6CNP
 
 | |
6GFJ
 
 | | Structure of RIP2 CARD domain fused to crystallisable MBP tag | | Descriptor: | Sugar ABC transporter substrate-binding protein,Receptor-interacting serine/threonine-protein kinase 2, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2018-04-30 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
6GHT
 
 | | HtxB D206A protein variant from Pseudomonas stutzeri in complex with hypophosphite to 1.12 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2018-05-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6CJ8
 
 | | CSP1 | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-02-26 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6CVR
 
 | | Human Aprataxin (Aptx) S242N bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6CVQ
 
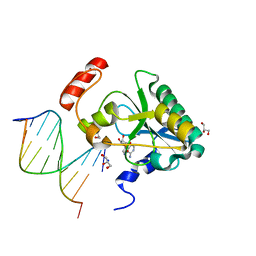 | | Human Aprataxin (Aptx) H201Q bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6C1I
 
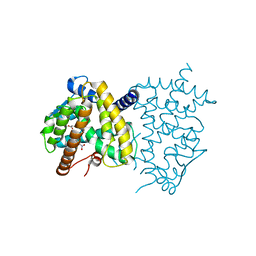 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with T0070907 | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Fuhrmann, J, Brust, R, Kojetin, D.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A structural mechanism for directing corepressor-selective inverse agonism of PPAR gamma.
Nat Commun, 9, 2018
|
|
6BPS
 
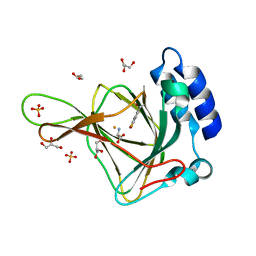 | | Crystal structure of cysteine-bound ferrous form of the uncrosslinked F2-Tyr157 human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6BPV
 
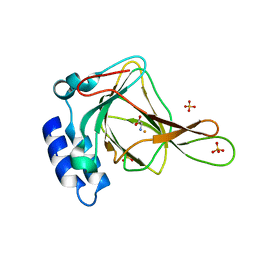 | | Crystal structure of cysteine-bound ferrous form of the matured F2-Tyr157 human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6BGF
 
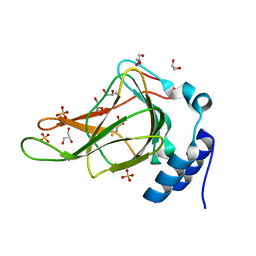 | | Crystal structure of cysteine-bound ferrous form of the crosslinked human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat.Chem.Biol., 14, 2018
|
|
6BZK
 
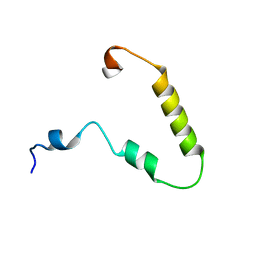 | | Solution structure of KTI55 | | Descriptor: | M protein | | Authors: | Castellino, F.J, Qiu, C, Yuan, Y. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6CMQ
 
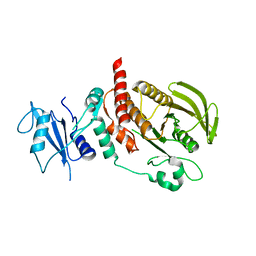 | | Structure of human SHP2 without N-SH2 domain | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
6COU
 
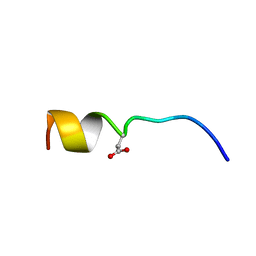 | | CSP2-E1Ad10 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6FPQ
 
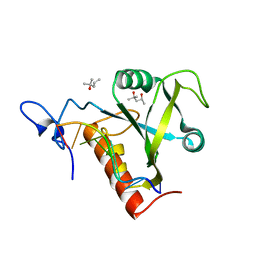 | | Structure of S. pombe Mmi1 in complex with 7-mer RNA | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, RNA (5'-R(*UP*UP*AP*AP*AP*CP*C)-3'), YTH domain-containing protein mmi1 | | Authors: | Stowell, J.A.W, Hill, C.H, Yu, M, Wagstaff, J.L, McLaughlin, S.H, Freund, S.M.V, Passmore, L.A. | | Deposit date: | 2018-02-11 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A low-complexity region in the YTH domain protein Mmi1 enhances RNA binding.
J. Biol. Chem., 293, 2018
|
|
6BPT
 
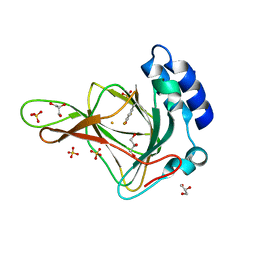 | | Crystal structure of ferrous form of the uncrosslinked F2-Tyr157 human cysteine dioxygenase | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6BYM
 
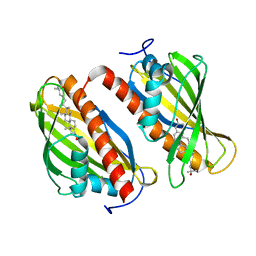 | | Crystal structure of the sterol-bound second StART domain of yeast Lam4 | | Descriptor: | 25-HYDROXYCHOLESTEROL, Sterol-binding protein | | Authors: | Jentsch, J.A, Kiburu, I.N, Wu, J, Pandey, K, Boudker, O, Menon, A.K. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of sterol binding and transport by a yeast StARkin domain.
J. Biol. Chem., 293, 2018
|
|
6BPW
 
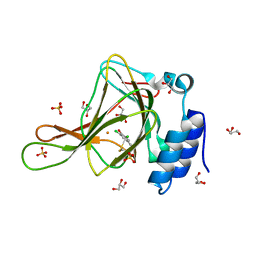 | | Crystal structure of ferrous form of the Cl2-Tyr157 human cysteine dioxygenase with both uncrosslinked and crosslinked cofactor | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6GFY
 
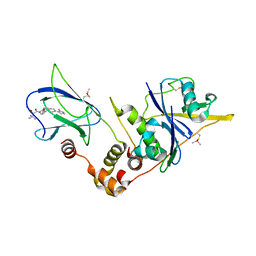 | | pVHL:EloB:EloC in complex with modified VH032 containing (3R,4S)-3-fluoro-4-hydroxyproline (ligand 14a) | | Descriptor: | (2~{R},3~{R},4~{S})-1-[(2~{S})-2-acetamido-3,3-dimethyl-butanoyl]-3-fluoranyl-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
6GOA
 
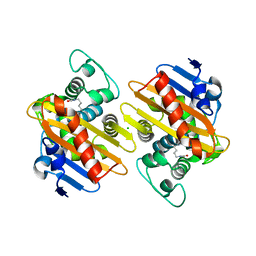 | |
6GXV
 
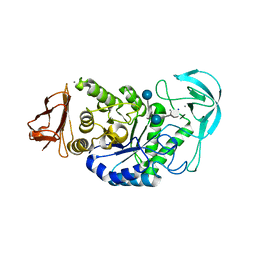 | | Amylase in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, A-amylase, CALCIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6GYA
 
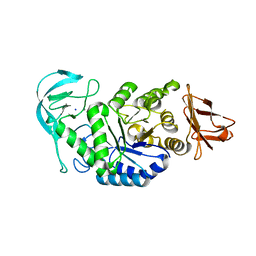 | | Amylase in complex with branched ligand | | Descriptor: | A-amylase, CALCIUM ION, SODIUM ION, ... | | Authors: | Agirre, J, Moroz, O, Meier, S, Brask, J, Munch, A, Hoff, T, Andersen, C, Wilson, K.S, Davies, G.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The structure of the AliC GH13 alpha-amylase from Alicyclobacillus sp. reveals the accommodation of starch branching points in the alpha-amylase family.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
