3HI6
 
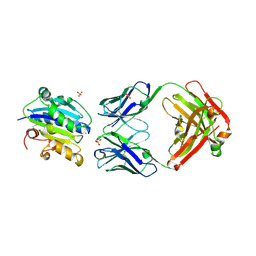 | |
4JV4
 
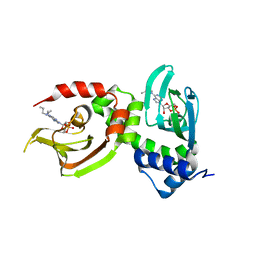 | | Crystal Structure of RIalpha(91-379) bound to HE33, a N6 di-propyl substituted cAMP analog | | Descriptor: | (2R,4aR,6R,7R,7aS)-6-[6-(dipropylamino)-9H-purin-9-yl]tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Brown, S.H.J, Cheng, C.Y, Saldanha, A.S, Wu, J, Cottam, H, Sankaran, B, Taylor, S.S. | | Deposit date: | 2013-03-25 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Implementing Fluorescence Anisotropy Screening and Crystallographic Analysis to Define PKA Isoform-Selective Activation by cAMP Analogs.
Acs Chem.Biol., 8, 2013
|
|
3VRE
 
 | | The crystal structure of hemoglobin from woolly mammoth in the deoxy form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3VRG
 
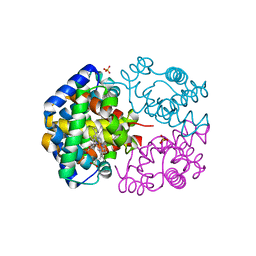 | | The crystal structure of hemoglobin from woolly mammoth in the met form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta/delta hybrid, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Noguchi, H, Campbell, K.L, Ho, C, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-04-09 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of haemoglobin from woolly mammoth in liganded and unliganded states.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FJR
 
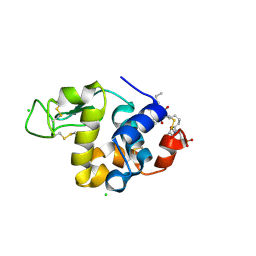 | | Mode of interaction of Merocyanine 540 with HEW Lysozyme | | Descriptor: | 3-[(2E)-2-[(2Z)-4-(1,3-dibutyl-4,6-dioxo-2-thioxotetrahydropyrimidin-5(2H)-ylidene)but-2-en-1-ylidene]-1,3-benzoxazol-3(2H)-yl]propane-1-sulfonic acid, CHLORIDE ION, Lysozyme C | | Authors: | Mitra, P, Banerjee, M, Basu, S, Biswas, S. | | Deposit date: | 2012-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mode of interaction of Merocyanine 540 dye with lysozyme
To be Published
|
|
3HEP
 
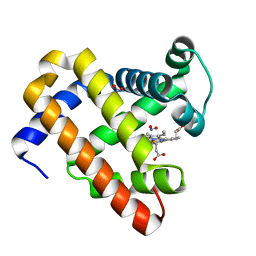 | | Ferric Horse Heart Myoglobin; H64V Mutant, Nitrite Modified | | Descriptor: | Myoglobin, NITRITE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yi, J, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2009-05-09 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The distal pocket histidine residue in horse heart myoglobin directs the o-binding mode of nitrite to the heme iron.
J.Am.Chem.Soc., 131, 2009
|
|
4FON
 
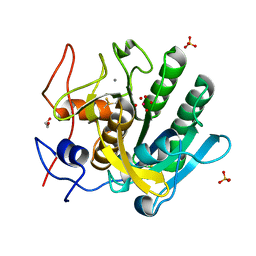 | |
4J68
 
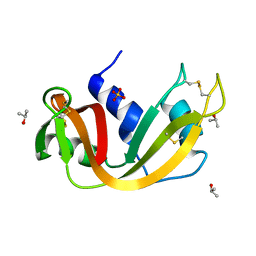 | |
4J6C
 
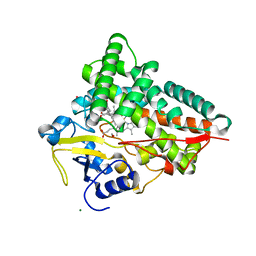 | |
3HG6
 
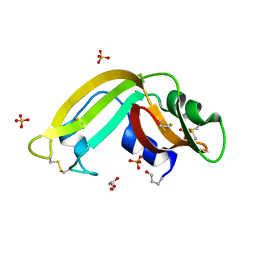 | | Crystal Structure of the Recombinant Onconase from Rana pipiens | | Descriptor: | GLYCEROL, Onconase, SULFATE ION | | Authors: | Camara-Artigas, A, Gavira, J.A, Casares-Atienza, S, Weininger, U, Balbach, J, Garcia-Mira, M.M. | | Deposit date: | 2009-05-13 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-state thermal unfolding of onconase.
Biophys.Chem., 159, 2011
|
|
3V9M
 
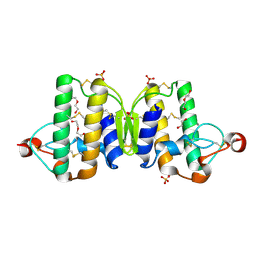 | | Phospholipase ACII4 from Australian King Brown Snake | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Guddat, L.W, Millers, E.K. | | Deposit date: | 2011-12-27 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (1.563 Å) | | Cite: | Mechanistic studies on the anticoagulant activity of a phospholipase A2 from the venom of the Australian King Brown Snake (Pseudechis australis)
To be Published
|
|
4J9F
 
 | |
4J9I
 
 | |
3VAL
 
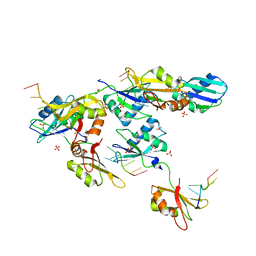 | | Structure of U2AF65 variant with BrU5C1 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*C*UP*UP*UP*(BRU)P*UP*U)-3'), SULFATE ION, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
4JA4
 
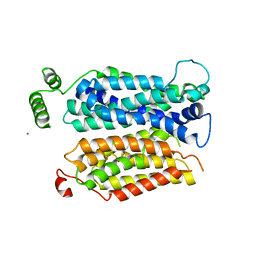 | | Inward open conformation of the xylose transporter XylE from E. coli | | Descriptor: | CADMIUM ION, D-xylose-proton symporter | | Authors: | Quistgaard, E.M, Low, C, Moberg, P, Tresaugues, L, Nordlund, P. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4FRJ
 
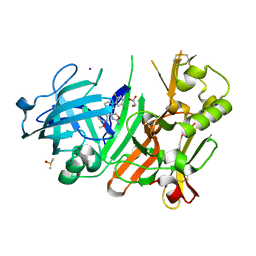 | | Crystal structure of BACE1 in complex with aminooxazoline xanthene 9l | | Descriptor: | (4S)-2'-(5-chloro-2-fluorophenyl)-7'-methoxyspiro[1,3-oxazole-4,9'-xanthen]-2-amine, Beta-secretase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-12 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure- and Property-Based Design of Aminooxazoline Xanthenes as Selective, Orally Efficacious, and CNS Penetrable BACE Inhibitors for the Treatment of Alzheimer's Disease.
J.Med.Chem., 55, 2012
|
|
4FRS
 
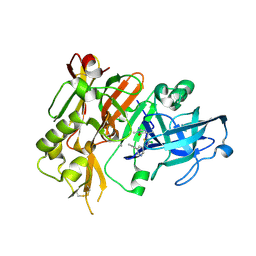 | |
4JBT
 
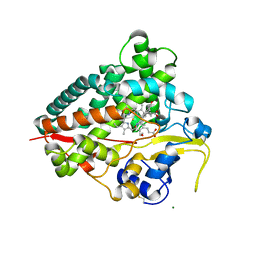 | |
3VDJ
 
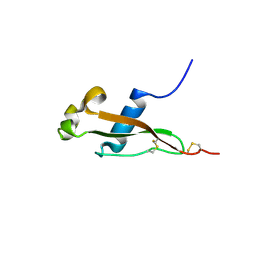 | | Crystal structure of circumsporozoite protein aTSR domain, R32 native form | | Descriptor: | Circumsporozoite (CS) protein | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VF5
 
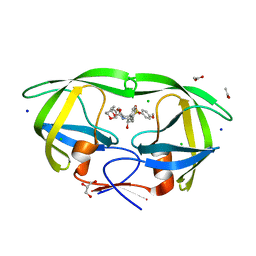 | | Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
4JEC
 
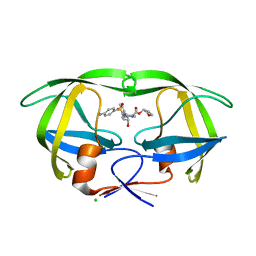 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | Deposit date: | 2013-02-26 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
3VHB
 
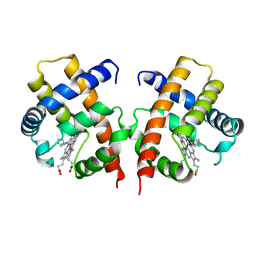 | | IMIDAZOLE ADDUCT OF THE BACTERIAL HEMOGLOBIN FROM VITREOSCILLA SP. | | Descriptor: | IMIDAZOLE, PROTEIN (HEMOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Boffi, A, Coletta, M, Mozzarelli, A, Pesce, A, Tarricone, C, Ascenzi, P. | | Deposit date: | 1999-03-17 | | Release date: | 1999-08-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anticooperative ligand binding properties of recombinant ferric Vitreoscilla homodimeric hemoglobin: a thermodynamic, kinetic and X-ray crystallographic study.
J.Mol.Biol., 291, 1999
|
|
4JK6
 
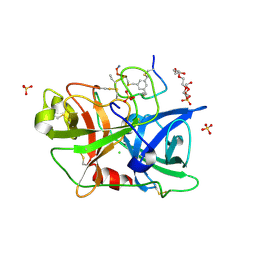 | | Human urokinase-type Plasminogen Activator (uPA) in complex with a bicyclic peptide inhibitor (UK18-D-Aba) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, CHLORIDE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Buth, S.A, Leiman, P.G, Chen, S, Heinis, C. | | Deposit date: | 2013-03-09 | | Release date: | 2013-07-17 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improving binding affinity and stability of Peptide ligands by substituting glycines with d-amino acids.
Chembiochem, 14, 2013
|
|
3HZC
 
 | |
3VWW
 
 | |
