4KWI
 
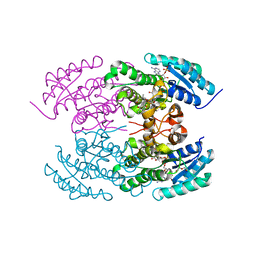 | | The crystal structure of angucycline C-6 ketoreductase LanV with bound NADP and 11-deoxy-6-oxylandomycinone | | Descriptor: | 1,8-dihydroxy-3-methyltetraphene-6,7,12(5H)-trione, DI(HYDROXYETHYL)ETHER, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Paananen, P, Patrikainen, P, Mantsala, P, Niemi, J, Niiranen, L, Metsa-Ketela, M. | | Deposit date: | 2013-05-24 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.
Biochemistry, 52, 2013
|
|
8GMS
 
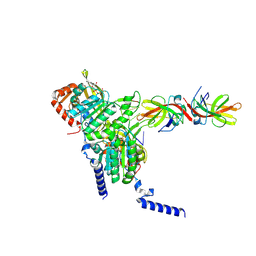 | | Structure of LexA in complex with RecA filament | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), LexA repressor, MAGNESIUM ION, ... | | Authors: | Gao, B, Feng, Y. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for regulation of SOS response in bacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4XT3
 
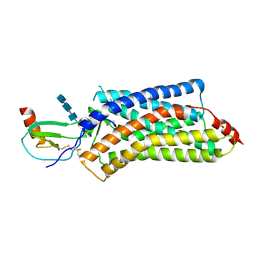 | | Structure of a viral GPCR bound to human chemokine CX3CL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Burg, J.S, Jude, K.M, Waghray, D, Garcia, K.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural biology. Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor.
Science, 347, 2015
|
|
5BRO
 
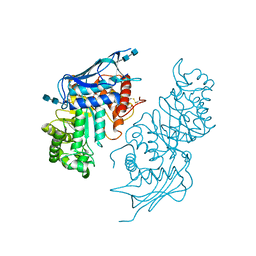 | | Crystal structure of modified HexB (modB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase subunit beta, FORMIC ACID, ... | | Authors: | Kitakaze, K, Maita, N, Itoh, K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protease-resistant modified human beta-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model.
J.Clin.Invest., 126, 2016
|
|
6VA8
 
 | |
5BCA
 
 | | BETA-AMYLASE FROM BACILLUS CEREUS VAR. MYCOIDES | | Descriptor: | CALCIUM ION, PROTEIN (1,4-ALPHA-D-GLUCAN MALTOHYDROLASE.) | | Authors: | Oyama, T, Kusunoki, M, Kishimoto, Y, Takasaki, Y, Nitta, Y. | | Deposit date: | 1999-03-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-amylase from Bacillus cereus var. mycoides at 2.2 A resolution.
J.Biochem.(Tokyo), 125, 1999
|
|
6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6BFS
 
 | | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies | | Descriptor: | Fab Heavy Chain, Fab light Chain, Granulocyte-macrophage colony-stimulating factor | | Authors: | Dhagat, U, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2017-10-26 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies suggests novel therapeutic opportunities.
MAbs, 10, 2018
|
|
6VDI
 
 | |
4XR7
 
 | | Structure of the Saccharomyces cerevisiae PAN2-PAN3 core complex | | Descriptor: | PAB-dependent poly(A)-specific ribonuclease subunit PAN2, PAB-dependent poly(A)-specific ribonuclease subunit PAN3 | | Authors: | Schafer, I.B, Rode, M, Bonneau, F, Schussler, S, Conti, E. | | Deposit date: | 2015-01-20 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.796 Å) | | Cite: | The structure of the Pan2-Pan3 core complex reveals cross-talk between deadenylase and pseudokinase.
Nat. Struct. Mol. Biol., 21, 2014
|
|
8GUE
 
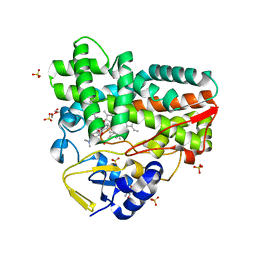 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-09-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
5BV5
 
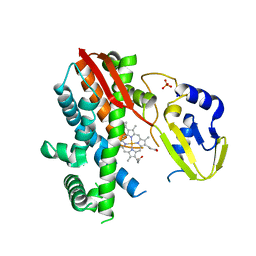 | | Structure of CYP119 with T213A and C317H mutations | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 119, PHOSPHATE ION, ... | | Authors: | Buller, A.R, Heel, T, McIntosh, J.A, Arnold, F.H. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Adaptability Facilitates Histidine Heme Ligation in a Cytochrome P450.
J.Am.Chem.Soc., 137, 2015
|
|
4LB9
 
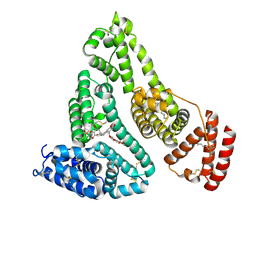 | | X-ray study of human serum albumin complexed with etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
5BU9
 
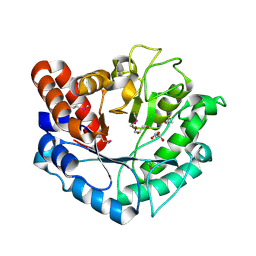 | | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333 | | Descriptor: | Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Chang, C, Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-03 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Crystal structure of Beta-N-acetylhexosaminidase from Beutenbergia cavernae DSM 12333
To Be Published
|
|
4Q0J
 
 | | Deinococcus radiodurans BphP photosensory module | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.747 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
4Q0H
 
 | | Deinococcus radiodurans BphP PAS-GAF | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.157 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
6UUT
 
 | |
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
6V15
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, G08 TCR alpha chain, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
2ZB3
 
 | | Crystal structure of mouse 15-ketoprostaglandin delta-13-reductase in complex with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
8JAR
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAL
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6UX0
 
 | | Isavuconazole bound complex of Acanthamoeba castellanii CYP51 | | Descriptor: | 4-{2-[(2R,3R)-3-(2,5-difluorophenyl)-3-hydroxy-4-(1H-1,2,4-triazol-1-yl)butan-2-yl]-1,3-thiazol-4-yl}benzonitrile, FE (III) ION, Obtusifoliol 14alphademethylase, ... | | Authors: | Sharma, V, Podust, L.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Domain-Swap Dimerization of Acanthamoeba castellanii CYP51 and a Unique Mechanism of Inactivation by Isavuconazole.
Mol.Pharmacol., 98, 2020
|
|
5BOO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Phillips, M, Deng, X, Tomchick, D. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
6UY4
 
 | | Crystal structure of dihydroorotate dehydrogenase from Schistosoma mansoni | | Descriptor: | 2-[(4-fluorophenyl)amino]-3-hydroxynaphthalene-1,4-dione, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Mori, R.M, Zapata, L.C.C, Nonato, M.C. | | Deposit date: | 2019-11-11 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structural basis for the function and inhibition of dihydroorotate dehydrogenase from Schistosoma mansoni.
Febs J., 288, 2021
|
|
