7LRC
 
 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
5U52
 
 | | 2 helix minimized version of the B-domain from Protein A (Z34C0 bound to IgG1 Fc (monoclinic form) | | Descriptor: | IGG1 FC, Mini Z domain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ultsch, M.H, Eigenbrot, C. | | Deposit date: | 2016-12-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | 3-2-1: Structural insights from stepwise shrinkage of a three-helix Fc-binding domain to a single helix.
Protein Eng. Des. Sel., 30, 2017
|
|
6CYY
 
 | | Mycobacterium tuberculosis transcriptional regulator | | Descriptor: | COENZYME A, HTH-type transcriptional regulator PrpR, IRON/SULFUR CLUSTER | | Authors: | Tang, S, Sacchettini, J. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structural and functional insight into the Mycobacterium tuberculosis protein PrpR reveals a novel type of transcription factor.
Nucleic Acids Res., 47, 2019
|
|
3HPV
 
 | |
1RIW
 
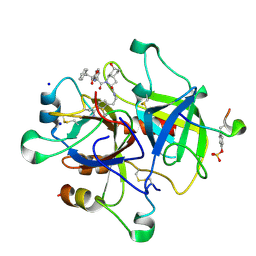 | | Thrombin in complex with natural product inhibitor Oscillarin | | Descriptor: | (2R,3AS,6R,7AS)-N-(2-{1-[AMINO(IMINO)METHYL]-2,5-DIHYDRO-1H-PYRROL-3-YL}ETHYL)-6-HYDROXY-1-{N-[(2S)-2-HYDROXY-3-PHENYLPROPANOYL]PHENYLALANYL}OCTAHYDRO-1H-INDOLE-2-CARBOXAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin IIB, ... | | Authors: | Hanessian, S, Tremblay, M, Petersen, J.F.W. | | Deposit date: | 2003-11-18 | | Release date: | 2004-11-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The N-acyloxyiminium ion aza-Prins route to octahydroindoles: total synthesis and structural confirmation of the antithrombotic marine natural product oscillarin
J.Am.Chem.Soc., 126, 2004
|
|
5UQC
 
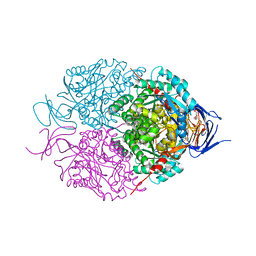 | | Crystal structure of mouse CRMP2 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Dihydropyrimidinase-related protein 2 | | Authors: | Khanna, M, Khanna, R, Perez-Miller, S, Francois-Moutal, L. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function.
Channels (Austin), 11, 2017
|
|
2PTQ
 
 | |
6GGU
 
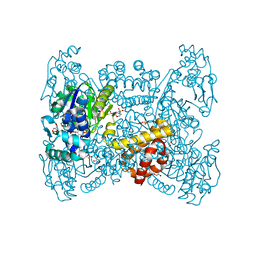 | | Crystal structure of native FE-hydrogenase from Methanothermobacter marburgensis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5,10-methenyltetrahydromethanopterin hydrogenase, GLYCEROL, ... | | Authors: | Wagner, T, Huang, G, Ermler, U, Shima, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Self-protection of the catalytic iron center of a methanogenic [Fe]-hydrogenase via a dynamic dimer-to-hexamer transformation
To Be Published
|
|
3ZHO
 
 | | X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVOPROTEIN WRBA | | Authors: | Kishko, I, Lapkouski, M, Brynda, J, Kuty, M, Carey, J, Smatanova, I.K, Ettrich, R. | | Deposit date: | 2012-12-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 A Resolution Crystal Structure of Escherichia Coli Wrba Holoprotein
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2PUW
 
 | | The crystal structure of isomerase domain of glucosamine-6-phosphate synthase from Candida albicans | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, isomerase domain of glutamine-fructose-6-phosphate transaminase (isomerizing) | | Authors: | Raczynska, J, Olchowy, J, Milewski, S, Rypniewski, W. | | Deposit date: | 2007-05-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.151 Å) | | Cite: | The Crystal and Solution Studies of Glucosamine-6-phosphate Synthase from Candida albicans
J.Mol.Biol., 372, 2007
|
|
7JLT
 
 | | Crystal Structure of SARS-CoV-2 NSP7-NSP8 complex. | | Descriptor: | Non-structural protein 7, Non-structural protein 8 | | Authors: | Biswal, M, Hai, R, Song, J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two conserved oligomer interfaces of NSP7 and NSP8 underpin the dynamic assembly of SARS-CoV-2 RdRP.
Nucleic Acids Res., 49, 2021
|
|
1YZV
 
 | |
6BZH
 
 | | Structure of mouse RIG-I tandem CARDs | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Babon, J.J, Nicholson, S.E. | | Deposit date: | 2017-12-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a second binding site on the TRIM25 B30.2 domain.
Biochem. J., 475, 2018
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5SZ8
 
 | |
7MC0
 
 | |
2PTS
 
 | |
4AIR
 
 | | Leishmania major cysteine synthase | | Descriptor: | CHLORIDE ION, Cysteine synthase, FGA-FGA-FGA-FGA, ... | | Authors: | Fyfe, P.K, Westrop, G.D, Coombs, G.H, Hunter, W.N. | | Deposit date: | 2012-02-13 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Leishmania Major Cysteine Synthase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
5TUG
 
 | |
4AUM
 
 | | Crystal structure, recombinant expression and mutagenesis studies of the bifunctional catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Yuzugullu, Y, Trinh, C.H, Smith, M.A, Pearson, A.R, Phillips, S.E.V, Sutay Kocabas, D, Bakir, U, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-05-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Recombinant Expression and Mutagenesis Studies of the Catalase with Oxidase Activity from Scytalidium Thermophilum
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5AKO
 
 | |
3IBX
 
 | | Crystal structure of F47Y variant of TenA (HP1287) from Helicobacter pylori | | Descriptor: | Putative thiaminase II | | Authors: | Barison, N, Cendron, L, Trento, A, Angelini, A, Zanotti, G. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and mutational analysis of TenA protein (HP1287) from the Helicobacter pylori thiamin salvage pathway - evidence of a different substrate specificity.
Febs J., 276, 2009
|
|
3IEY
 
 | |
1Y3I
 
 | | Crystal Structure of Mycobacterium tuberculosis NAD kinase-NAD complex | | Descriptor: | GLYCEROL, Inorganic polyphosphate/ATP-NAD kinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mori, S, Yamasaki, M, Maruyama, Y, Momma, K, Kawai, S, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-25 | | Release date: | 2005-01-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex
Biochem.Biophys.Res.Commun., 327, 2005
|
|
3HQ0
 
 | | Crystal Structure Analysis of the 2,3-dioxygenase LapB from Pseudomonas in complex with a product | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohepta-2,4-dienoic acid, Catechol 2,3-dioxygenase, FE (III) ION | | Authors: | Cho, J.-H, Rhee, S. | | Deposit date: | 2009-06-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and functional analysis of the extradiol dioxygenase LapB from a long-chain alkylphenol degradation pathway in Pseudomonas
J.Biol.Chem., 284, 2009
|
|
