6YPG
 
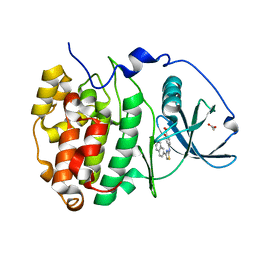 | |
6YPK
 
 | | Crystal Structure of CK2alpha with GTP bound | | Descriptor: | Casein kinase II subunit alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Proposed Allosteric Inhibitors Bind to the ATP Site of CK2 alpha.
J.Med.Chem., 63, 2020
|
|
1G3F
 
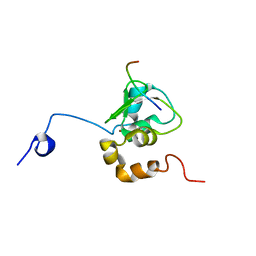 | | NMR STRUCTURE OF A 9 RESIDUE PEPTIDE FROM SMAC/DIABLO COMPLEXED TO THE BIR3 DOMAIN OF XIAP | | Descriptor: | INHIBITOR OF APOPTOSIS PROTEIN 3, SMAC, ZINC ION | | Authors: | Liu, Z, Sun, C, Olejniczak, E.T, Meadows, R.P, Betz, S.F, Oost, T, Herrmann, J, Wu, J.C, Fesik, S.W. | | Deposit date: | 2000-10-24 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain.
Nature, 408, 2000
|
|
6YQ2
 
 | |
6YSB
 
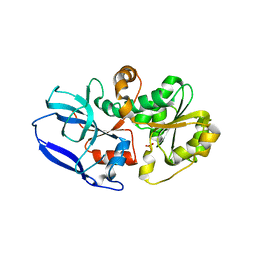 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) apo form | | Descriptor: | 2-alkenal reductase (NADP(+)-dependent)-like, SULFATE ION | | Authors: | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6Y9W
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,8) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8J56
 
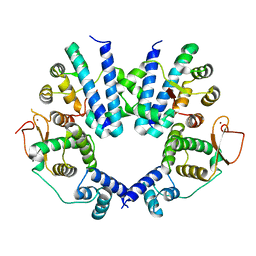 | | Crystal structure of the FlhDC complex from Cupriavidus necator | | Descriptor: | Flagellar transcriptional regulator FlhC, Flagellar transcriptional regulator FlhD, ZINC ION | | Authors: | Cho, S.Y, Oh, H.B, Yoon, S.I. | | Deposit date: | 2023-04-21 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hexameric structure of the flagellar master regulator FlhDC from Cupriavidus necator and its interaction with flagellar promoter DNA.
Biochem.Biophys.Res.Commun., 672, 2023
|
|
6YBF
 
 | |
4HEA
 
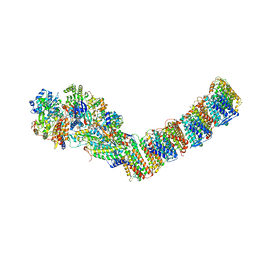 | | Crystal structure of the entire respiratory complex I from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Baradaran, R, Berrisford, J.M, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3027 Å) | | Cite: | Crystal structure of the entire respiratory complex I.
Nature, 494, 2013
|
|
8FAA
 
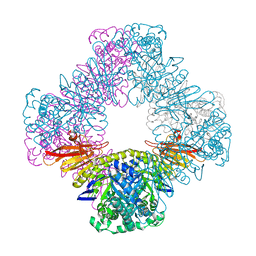 | |
6YEI
 
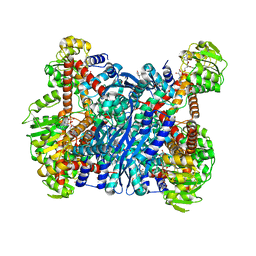 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in complex with NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
5CLC
 
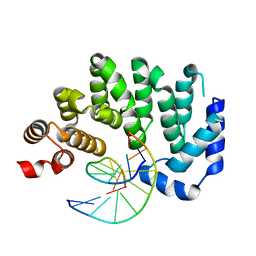 | |
6YE2
 
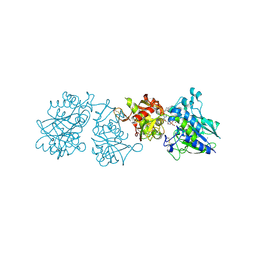 | |
6YOD
 
 | | Structure of Lysozyme from SiN IMISX setup collected by rotation serial crystallography on crystals prelocated by 2D X-ray phase-contrast imaging | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-(2-METHOXYETHOXY)ETHANOL, ACETIC ACID, ... | | Authors: | Huang, C.-Y, Martiel, I, Villanueva-Perez, P, Panepucci, E, Caffrey, M, Wang, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Low-dose in situ prelocation of protein microcrystals by 2D X-ray phase-contrast imaging for serial crystallography.
Iucrj, 7, 2020
|
|
1G9L
 
 | | SOLUTION STRUCTURE OF THE PABC DOMAIN OF HUMAN POLY(A) BINDING PROTEIN | | Descriptor: | POLYADENYLATE-BINDING PROTEIN 1 | | Authors: | Kozlov, G, Trempe, J.-F, Khaleghpour, K, Kahvejian, A, Ekiel, I, Gehring, K. | | Deposit date: | 2000-11-24 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the C-terminal PABC domain of human poly(A)-binding protein.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8FA5
 
 | |
5N1J
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Peptidoglycan N-acetylglucosamine deacetylase, ... | | Authors: | Giastas, P, Andreou, A, Balomenou, S, Bouriotis, V, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
5MYY
 
 | | Hen Egg-White Lysozyme (HEWL) cocrystallized in the presence of Cadmium sulphate | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Panneerselvam, S, Burkhardt, A, Meents, A. | | Deposit date: | 2017-01-30 | | Release date: | 2017-07-12 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rapid cadmium SAD phasing at the standard wavelength (1 angstrom ).
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5MZ8
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
6YPY
 
 | |
6YOW
 
 | |
6YOZ
 
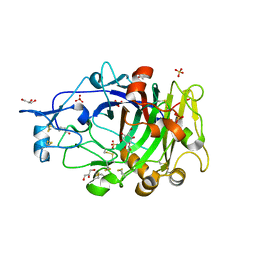 | | HiCel7B labelled with b-1,4-glucosyl cyclophellitol | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Glycosylated cyclophellitol-derived activity-based probes and inhibitors for cellulases.
Rsc Chem Biol, 1, 2020
|
|
1W0F
 
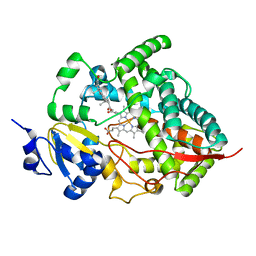 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
6YPA
 
 | |
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
