5FPO
 
 | | Structure of Bacterial DNA Ligase with small-molecule ligand 1H- indazol-7-amine (AT4213) in an alternate binding site. | | Descriptor: | 1H-indazol-7-amine, DNA LIGASE | | Authors: | Jhoti, H, Ludlow, R.F, Pathuri, P, Saini, H.K, Tickle, I.J, Tisi, D, Verdonk, M, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4MKY
 
 | |
5V1J
 
 | | DNA polymerase beta open product complex with 8-oxoG:C and inserted dCTP | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG)P*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Smith, M.R, Whitaker, A.M, Schaich, M.A. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
5V1G
 
 | | DNA polymerase beta binary complex with 8-oxoG at the primer terminus | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Smith, M.R, Whitaker, A.M, Schaich, M.A. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
5V1H
 
 | | DNA polymerase beta binary complex with 8-oxoG:A at the primer terminus | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Freudenthal, B.D, Schaich, M.A, Whitaker, A.M, Smith, M.R. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Capturing a mammalian DNA polymerase extending from an oxidized nucleotide.
Nucleic Acids Res., 45, 2017
|
|
6ZHE
 
 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2R9L
 
 | | Polymerase Domain from Mycobacterium tuberculosis Ligase D in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*DGP*DCP*DCP*DGP*DCP*DAP*DAP*DCP*DGP*DCP*DA)-3'), ... | | Authors: | Brissett, N.C, Fox, G.C, Pitcher, R.S, Doherty, A.J. | | Deposit date: | 2007-09-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a NHEJ polymerase-mediated DNA synaptic complex
Science, 318, 2007
|
|
7Z88
 
 | | DNA-PK in the intermediate state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z87
 
 | | DNA-PK in the active state | | Descriptor: | (~{S})-[2-chloranyl-4-fluoranyl-5-(7-morpholin-4-ylquinazolin-4-yl)phenyl]-(6-methoxypyridazin-3-yl)methanol, DNA (26-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Liang, S, Blundell, T.L. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Human DNA-dependent protein kinase activation mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8UCH
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCE
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCI
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G + AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP dependent DNA ligase, GLYCEROL, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCF
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K238G | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
8UCG
 
 | | Thermophilic RNA Ligase from Palaeococcus pacificus K92A | | Descriptor: | ATP dependent DNA ligase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Rousseau, M.D, Hicks, J.L, Oulavallickal, T, Williamson, A, Arcus, V.L, Patrick, M.W. | | Deposit date: | 2023-09-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.
Nucleic Acids Res., 52, 2024
|
|
6WH2
 
 | | Structure of the C-terminal BRCT domain of human XRCC1 | | Descriptor: | X-ray repair cross complementing protein 1 variant | | Authors: | Pourfarjam, Y, Ellenberger, T, Tainer, J.A, Tomkinson, A.E, Kim, I.K. | | Deposit date: | 2020-04-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | An atypical BRCT-BRCT interaction with the XRCC1 scaffold protein compacts human DNA Ligase III alpha within a flexible DNA repair complex.
Nucleic Acids Res., 49, 2021
|
|
8P26
 
 | | Crystal structure of Arabidopsis thaliana PAXX | | Descriptor: | U2 small nuclear ribonucleoprotein auxiliary factor-like protein | | Authors: | Ochi, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plant PAXX has an XLF-like function and stimulates DNA end joining by the Ku-DNA ligase IV/XRCC4 complex.
Plant J., 116, 2023
|
|
7K11
 
 | |
7K10
 
 | |
7K19
 
 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex I) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1B
 
 | | CryoEM structure of DNA-PK catalytic subunit complexed with DNA (Complex II) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*AP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7OTP
 
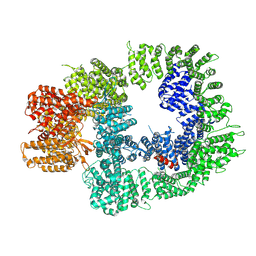 | | DNA-PKcs in complex with ATPgammaS-Mg | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Liang, S, Thomas, S.E, Blundell, T.L. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into inhibitor regulation of the DNA repair protein DNA-PKcs.
Nature, 601, 2022
|
|
5Y3R
 
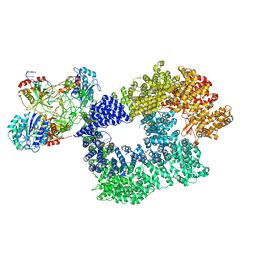 | | Cryo-EM structure of Human DNA-PK Holoenzyme | | Descriptor: | DNA (34-MER), DNA (36-MER), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Yin, X, Liu, M, Tian, Y, Wang, J, Xu, Y. | | Deposit date: | 2017-07-29 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-EM structure of human DNA-PK holoenzyme
Cell Res., 27, 2017
|
|
5W1R
 
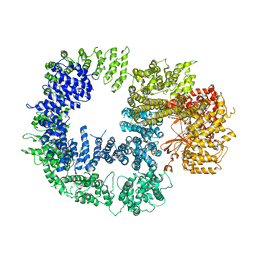 | | Cryo-EM structure of DNAPKcs | | Descriptor: | DNA-dependent protein kinase catalytic subunit | | Authors: | Sharif, H, Li, Y, Wu, H. | | Deposit date: | 2017-06-04 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the DNA-PK holoenzyme.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ZH2
 
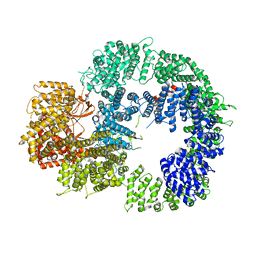 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZFP
 
 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
