3EOK
 
 | |
3EKV
 
 | |
3EL5
 
 | |
4I3D
 
 | | Crystal structure of fluorescent protein UnaG N57A mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bilirubin-inducible fluorescent protein UnaG | | Authors: | Kumagai, A, Ando, R, Miyatake, H, Miyawaki, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | A bilirubin-inducible fluorescent protein from eel muscle
Cell(Cambridge,Mass.), 153, 2013
|
|
4DQE
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, Aspartyl protease | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
4DFG
 
 | | Crystal Structure of Wild-type HIV-1 Protease with Cyclopentyltetrahydro- furanyl Urethanes as P2-ligand, GRL-0249A | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Substituent effects on P2-cyclopentyltetrahydrofuranyl urethanes: Design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3EM4
 
 | | Crystal structure of atazanavir (ATV) in complex with I50L/A71V drug-resistant HIV-1 protease | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N, Royer, C, Schiffer, C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Structural studies on atazanavir-specific I50L drug-resistant HIV-1 protease mutant
To be Published
|
|
4CN9
 
 | | structure of proximal thread matrix protein 1 (PTMP1) from the mussel byssus with zinc occupied MIDAS motif | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PROXIMAL THREAD MATRIX PROTEIN 1, ... | | Authors: | Gertz, M, Suhre, M.H, Scheibel, T, Steegborn, C. | | Deposit date: | 2014-01-21 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Features of a Collagen-Binding Matrix Protein from the Mussel Byssus.
Nat.Commun., 5, 2014
|
|
3E4N
 
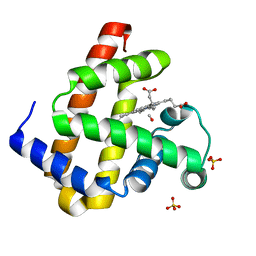 | | Carbonmonoxy Sperm Whale Myoglobin at 40 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3E5K
 
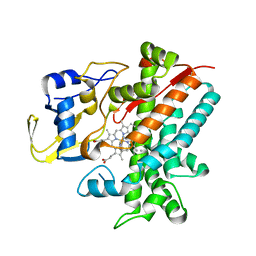 | | Crystal structure of CYP105P1 wild-type 4-phenylimidazole complex | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Cytochrome P450 (Cytochrome P450 hydroxylase), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, L.H, Fushinobu, S, Ikeda, H, Wakagi, T, Shoun, H. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine ligation state
J.Bacteriol., 191, 2009
|
|
3ZKQ
 
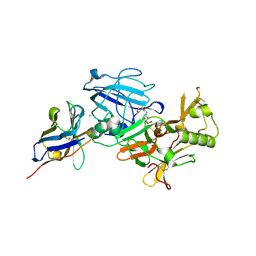 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3E6I
 
 | |
4I9S
 
 | |
4CW9
 
 | | Entamoeba histolytica thiredoxin C34S mutant | | Descriptor: | THIOREDOXIN | | Authors: | Parsonage, D, Kells, P.M, Vieira, D.F, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2014-04-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
4I8G
 
 | | Bovine trypsin at 0.8 resolution | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Dauter, Z, Liebschner, D, Dauter, M, Brzuszkiewicz, A. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-19 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | On the reproducibility of protein crystal structures: five atomic resolution structures of trypsin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3FE1
 
 | | Crystal structure of the human 70kDa heat shock protein 6 (Hsp70B') ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 6, ... | | Authors: | Wisniewska, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
3FFX
 
 | | Crystal Structure of CheY triple mutant F14E, N59R, E89H complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
3F7Q
 
 | |
3ZPI
 
 | | PikC D50N mutant in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, CYTOCHROME P450 HYDROXYLASE PIKC, FORMIC ACID, ... | | Authors: | Podust, L.M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Recognition of Synthetic Substrates by P450 Pikc
To be Published
|
|
4HZI
 
 | |
3ZCF
 
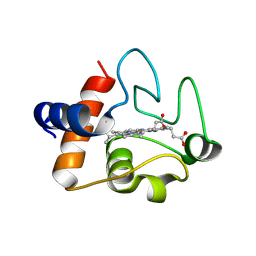 | | Structure of recombinant human cytochrome c | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Rajagopal, B.S, Worrall, J.A.R, Hough, M.A. | | Deposit date: | 2012-11-20 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Hydrogen Peroxide Induced Radical Behaviour in Human Cytochrome C Phospholipid Complexes: Implications for the Enhanced Pro-Apoptotic Activity of the G41S Mutant
Biochem.J., 456, 2013
|
|
4HZV
 
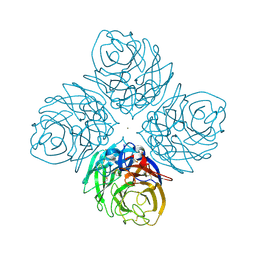 | | The crystal structure of influenza A neuraminidase N3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
4I0I
 
 | |
4CY7
 
 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form II | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
3ZI3
 
 | | Crystal structure of the B24His-insulin - human analogue | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Zakova, L, Kletvikova, E, Veverka, V, Lepsik, M, Watson, C.J, Turkenburg, J.P, Jiracek, J, Brzozowski, A.M. | | Deposit date: | 2013-01-02 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Integrity of the B24 Site in Human Insulin is Important for Hormone Functionality
J.Biol.Chem., 288, 2013
|
|
