2H5Q
 
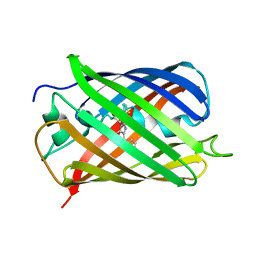 | | Crystal structure of mCherry | | Descriptor: | mCherry | | Authors: | Shu, X, Remington, S.J. | | Deposit date: | 2006-05-26 | | Release date: | 2006-08-22 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Novel Chromophores and Buried Charges Control Color in mFruits
Biochemistry, 45, 2006
|
|
3KK8
 
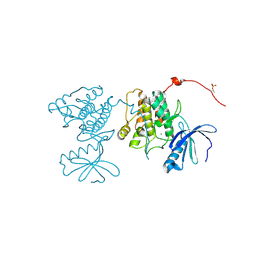 | | CaMKII Substrate Complex A | | Descriptor: | Calcium/calmodulin dependent protein kinase II, MAGNESIUM ION | | Authors: | Kuriyan, J, Chao, L.H, Pellicena, P, Deindl, S, Barclay, L.A, Schulman, H. | | Deposit date: | 2009-11-04 | | Release date: | 2010-02-09 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Intersubunit capture of regulatory segments is a component of cooperative CaMKII activation.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1ZDJ
 
 | | STRUCTURE OF BACTERIOPHAGE COAT PROTEIN-LOOP RNA COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*C)-3') | | Authors: | Grahn, E, Stonehouse, N, Valegard, K, Vandenworm, S, Liljas, L. | | Deposit date: | 1997-12-18 | | Release date: | 1998-07-08 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of RNA hairpins in complexes with recombinant MS2 capsids: implications for binding requirements.
RNA, 5, 1999
|
|
1SMY
 
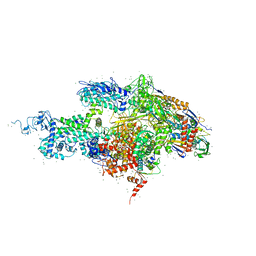 | | Structural basis for transcription regulation by alarmone ppGpp | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Patlan, V, Sekine, S, Vassylyeva, M.N, Hosaka, T, Ochi, K, Yokoyama, S, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-10 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for transcription regulation by alarmone ppGpp
Cell(Cambridge,Mass.), 117, 2004
|
|
1YVY
 
 | | Crystal structure of Anaerobiospirillum succiniciproducens phosphoenolpyruvate carboxykinase | | Descriptor: | Phosphoenolpyruvate carboxykinase [ATP] | | Authors: | Cotelesage, J.J, Prasad, L, Zeikus, J.G, Laivenieks, M, Delbaere, L.T. | | Deposit date: | 2005-02-16 | | Release date: | 2005-07-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Anaerobiospirillum succiniciproducens PEP carboxykinase reveals an important active site loop.
Int.J.Biochem.Cell Biol., 37, 2005
|
|
3KMQ
 
 | | G62S mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA, tetragonal structure | | Descriptor: | 3D polymerase, RNA (5'-R(*GP*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*CP*C)-3') | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
1S5N
 
 | | Xylose Isomerase in Substrate and Inhibitor Michaelis States: Atomic Resolution Studies of a Metal-Mediated Hydride Shift | | Descriptor: | HYDROXIDE ION, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Fenn, T.D, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Xylose isomerase in substrate and inhibitor michaelis States: atomic resolution studies of a metal-mediated hydride shift(,).
Biochemistry, 43, 2004
|
|
1SE8
 
 | | Structure of single-stranded DNA-binding protein (SSB) from D. radiodurans | | Descriptor: | Single-strand binding protein | | Authors: | Bernstein, D.A, Eggington, J.M, Killoran, M.P, Misic, A.M, Cox, M.M, Keck, J.L. | | Deposit date: | 2004-02-16 | | Release date: | 2004-06-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S48
 
 | | Crystal structure of RNA-dependent RNA polymerase construct 1 (residues 71-679) from BVDV | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1RXV
 
 | | Crystal Structure of A. Fulgidus FEN-1 bound to DNA | | Descriptor: | 5'-d(*T*pA*pG*pC*pA*pT*pC*pG*pG), Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
1S4F
 
 | | Crystal Structure of RNA-dependent RNA polymerase construct 2 from bovine viral diarrhea virus (BVDV) | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-16 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S06
 
 | | Crystal Structure of the R253K Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
3L3I
 
 | | Crystal structure of HLA-B*4402 in complex with the F7A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-17 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1S70
 
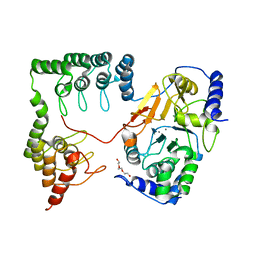 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
2FAI
 
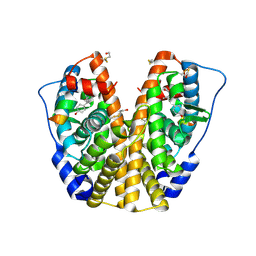 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-2M and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2S,5S,9R)-5-(HYDROXYMETHYL)-8,9-DIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
2BMH
 
 | |
3MK4
 
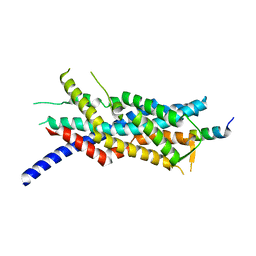 | | X-Ray structure of human PEX3 in complex with a PEX19 derived peptide | | Descriptor: | Peroxisomal biogenesis factor 19, Peroxisomal biogenesis factor 3 | | Authors: | Schmidt, F, Treiber, N, Dodt, G, Stehle, T. | | Deposit date: | 2010-04-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19
J.Biol.Chem., 285, 2010
|
|
3MP5
 
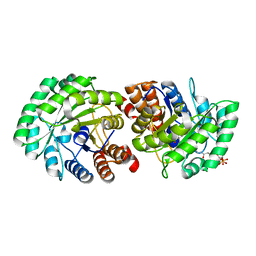 | | Crystal Structure of Human Lyase R41M in complex with HMG-CoA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, Hydroxymethylglutaryl-CoA lyase, MAGNESIUM ION | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
2GRV
 
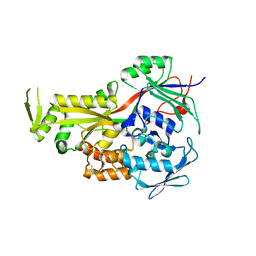 | | Crystal Structure of LpqW | | Descriptor: | LpqW | | Authors: | Marland, Z, Rossjohn, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-07-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hijacking of a Substrate-binding Protein Scaffold for use in Mycobacterial Cell Wall Biosynthesis
J.Mol.Biol., 359, 2006
|
|
3M5M
 
 | |
1PL5
 
 | | Crystal Structure Analysis of the Sir4p C-terminal Coiled Coil | | Descriptor: | Regulatory protein SIR4 | | Authors: | Murphy, G.A, Spedale, E.J, Powell, S.T, Pillus, L, Schultz, S.C, Chen, L. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Sir4 C-terminal coiled coil is required for telomeric and mating type silencing in Saccharomyces cerevisiae.
J.Mol.Biol., 334, 2003
|
|
3M4V
 
 | | Crystal structure of the A330P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-03-12 | | Release date: | 2011-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the properties of two single-site proline mutants of CYP102A1 (P450BM3)
Chembiochem, 11, 2010
|
|
2ALP
 
 | | REFINED STRUCTURE OF ALPHA-LYTIC PROTEASE AT 1.7 ANGSTROMS RESOLUTION. ANALYSIS OF HYDROGEN BONDING AND SOLVENT STRUCTURE | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Fujinaga, M, Delbaere, L.T.J, Brayer, G.D, James, M.N.G. | | Deposit date: | 1985-03-07 | | Release date: | 1985-07-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure.
J.Mol.Biol., 184, 1985
|
|
2BE5
 
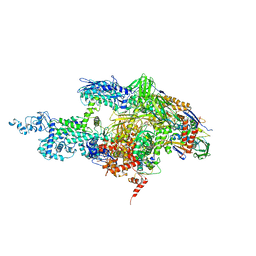 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with inhibitor tagetitoxin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Vassylyev, D.G, Svetlov, V, Vassylyeva, M.N, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Artsimovitch, I, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-22 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for transcription inhibition by tagetitoxin
Nat.Struct.Mol.Biol., 12, 2005
|
|
3MGQ
 
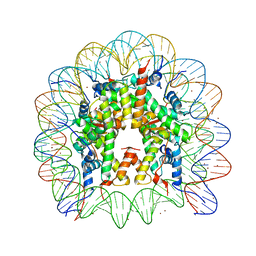 | | Binding of Nickel ions to the Nucleosome Core Particle | | Descriptor: | CHLORIDE ION, DNA (147-MER), Histone H2A, ... | | Authors: | Mohideen, K, Muhammad, R, Davey, C.A. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Perturbations in nucleosome structure from heavy metal association.
Nucleic Acids Res., 38, 2010
|
|
