5FDD
 
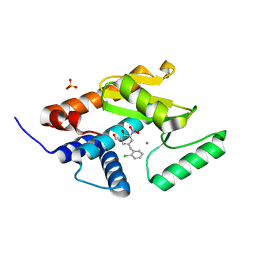 | | Endonuclease inhibitor 1 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 5-(2-chlorobenzyl)-2-hydroxy-3-nitrobenzaldehyde, MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2015-12-16 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4YBF
 
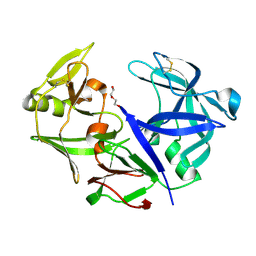 | | Aspartic Proteinase Sapp2 Secreted from Candida Parapsilosis at 1.25 A Resolution | | Descriptor: | Candidapepsin-2, DI(HYDROXYETHYL)ETHER | | Authors: | Dostal, J, Hruskova-Heidingsfeldova, O, Rezacova, P, Brynda, J, Mareckova, L, Pichova, I. | | Deposit date: | 2015-02-18 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6U2V
 
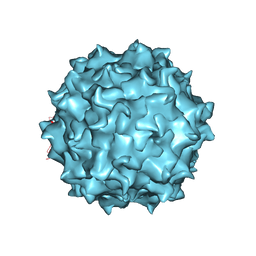 | | AAV8 Baculovirus-Sf9 produced, full capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
Mol Ther Methods Clin Dev, 18, 2020
|
|
6U3F
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Panton-Valentine Leucocidin F, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U3T
 
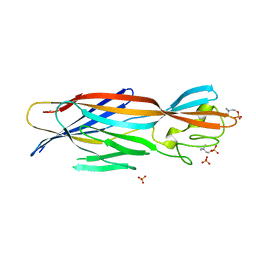 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
6U4L
 
 | | cysteine dioxygenase variant - C93E | | Descriptor: | ACETATE ION, Cysteine dioxygenase type 1, FE (III) ION | | Authors: | Meneely, K.M, Chilton, A.S, Forbes, D.L, Ellis, H.R, Lamb, A.L. | | Deposit date: | 2019-08-26 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | The 3-His Metal Coordination Site Promotes the Coupling of Oxygen Activation to Cysteine Oxidation in Cysteine Dioxygenase.
Biochemistry, 59, 2020
|
|
6U4P
 
 | | Structure-based discovery of a novel small-molecule inhibitor of methicillin-resistant S. aureus | | Descriptor: | Alpha-hemolysin, SULFATE ION, fos-choline-14 | | Authors: | Liu, J, Kozhaya, L, Torres, V.J, Unutmaz, D, Lu, M. | | Deposit date: | 2019-08-26 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based discovery of a small-molecule inhibitor of methicillin-resistantStaphylococcus aureusvirulence.
J.Biol.Chem., 295, 2020
|
|
2XXY
 
 | |
7YE7
 
 | | Crystal structure of SARS-CoV-2 soluble dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein, nonane | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
2XWR
 
 | |
7LJM
 
 | | Structure of the Salmonella enterica CD-NTase CdnD in complex with GTP | | Descriptor: | CD-NTase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.W, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
7LJL
 
 | | Structure of the Enterobacter cloacae CD-NTase CdnD in complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Cyclic AMP-AMP-GMP synthase, ... | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.W, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
7YE8
 
 | | Crystal structure of SARS-CoV-2 refolded dimeric ORF9b | | Descriptor: | N-OCTANE, ORF9b protein | | Authors: | Jin, X, Chai, Y, Qi, J, Song, H, Gao, G.F. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural characterization of SARS-CoV-2 dimeric ORF9b reveals potential fold-switching trigger mechanism.
Sci China Life Sci, 66, 2023
|
|
7XZZ
 
 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7LJO
 
 | | Structure of the Bacteroides fragilis CD-NTase CdnB in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CD-NTase, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.T, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
7LJN
 
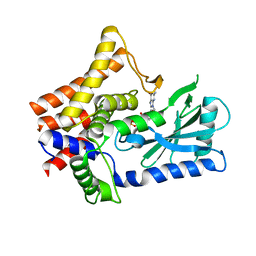 | | Structure of the Bradyrhizobium diazoefficiens CD-NTase CdnG in complex with GTP | | Descriptor: | CD-NTase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Govande, A, Lowey, B, Eaglesham, J.B, Whiteley, A.T, Kranzusch, P.J. | | Deposit date: | 2021-01-29 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of CD-NTase nucleotide selection in CBASS anti-phage defense.
Cell Rep, 35, 2021
|
|
4WY5
 
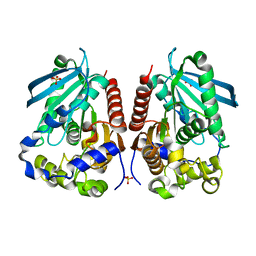 | | Structural analysis of two fungal esterases from Rhizomucor miehei explaining their substrate specificity | | Descriptor: | Esterase, SULFATE ION | | Authors: | Qin, Z, Yang, S, Duan, X, Yan, Q, Jiang, Z. | | Deposit date: | 2014-11-15 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the substrate specificity of two esterases from the thermophilic Rhizomucor miehei
J.Lipid Res., 56, 2015
|
|
7LH5
 
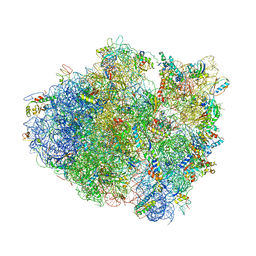 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with plazomicin, mRNA and tRNAs | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, 16S ribosomal RNA, 23S ribosomal RNA, ... | | Authors: | Golkar, T, Berghuis, A.M, Schmeing, T.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structural basis for plazomicin antibiotic action and resistance.
Commun Biol, 4, 2021
|
|
4X09
 
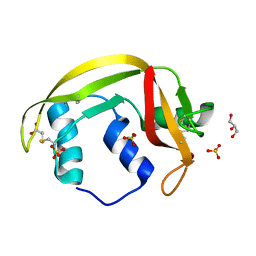 | | Structure of human RNase 6 in complex with sulphate anions | | Descriptor: | GLYCEROL, Ribonuclease K6, SULFATE ION | | Authors: | Prats-Ejarque, G, Arranz-Trullen, J, Blanco, J.A, Pulido, D, Moussaoui, M, Boix, E. | | Deposit date: | 2014-11-21 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | The first crystal structure of human RNase 6 reveals a novel substrate-binding and cleavage site arrangement.
Biochem.J., 473, 2016
|
|
7B2G
 
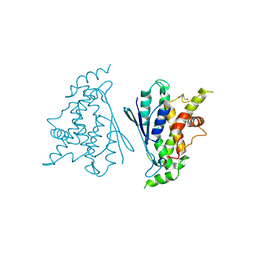 | | Crystal structure of R120Q GDAP1 mutant | | Descriptor: | GLYCEROL, Ganglioside-induced differentiation-associated protein 1 | | Authors: | Nguyen, G.T.T, Sutinen, A, Kursula, P. | | Deposit date: | 2020-11-26 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Conserved intramolecular networks in GDAP1 are closely connected to CMT-linked mutations and protein stability.
Plos One, 18, 2023
|
|
2VY8
 
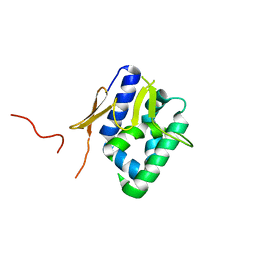 | | The 627-domain from influenza A virus polymerase PB2 subunit with Glu- 627 | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Crepin, T, Guilligay, D, Ruigrok, R, Cusack, S, Hart, D. | | Deposit date: | 2008-07-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase Pb2 Subunit
Plos Pathog., 4, 2008
|
|
5GTN
 
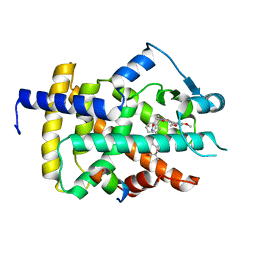 | | Human PPARgamma ligand binding dmain complexed with R35 | | Descriptor: | 2-[4-[5-[(1~{R})-1-[(3,5-dimethoxyphenyl)carbamoyl-(phenylmethyl)carbamoyl]oxypropyl]-1,2-oxazol-3-yl]phenoxy]-2-methyl-propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y, Suh, S.W. | | Deposit date: | 2016-08-22 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for differential activities of enantiomeric PPAR gamma agonists: Binding of S35 to the alternate site.
Biochim. Biophys. Acta, 1865, 2017
|
|
4H1O
 
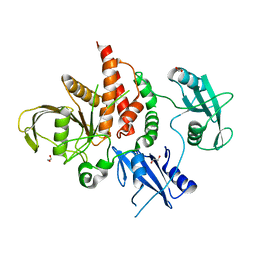 | | Crystal structure of the tyrosine phosphatase SHP-2 with D61G mutation | | Descriptor: | 1,2-ETHANEDIOL, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Qiu, W, Lin, A, Hutchinson, A, Romanov, V, Ruzanov, M, Thompson, C, Lam, K, Kisselman, G, Battaile, K, Chirgadze, N.Y. | | Deposit date: | 2012-09-11 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the tyrosine phosphatase SHP-2 with D61G mutation
To be Published
|
|
7ABR
 
 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
6U7H
 
 | | Cryo-EM structure of the HCoV-229E spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Z, Benlekbir, S, Rubinstein, J.L, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
