7ENC
 
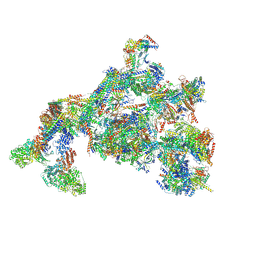 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
5SVA
 
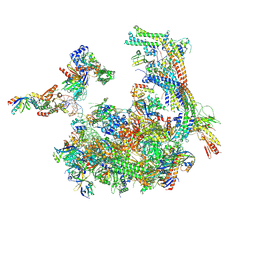 | | Mediator-RNA Polymerase II Pre-Initiation Complex | | Descriptor: | 108bp HIS4 Promoter Non-template Strand (-92/+16), 108bp HIS4 Promoter Template Strand (+16/-92), DNA repair helicase RAD25, ... | | Authors: | Robinson, P.J, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2016-08-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (15.3 Å) | | Cite: | Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex.
Cell, 166, 2016
|
|
7UBM
 
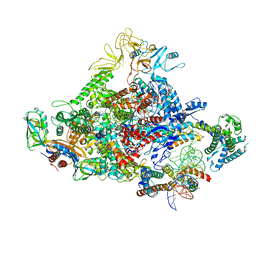 | |
4UIG
 
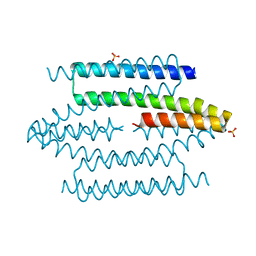 | |
7DOG
 
 | |
7SZK
 
 | | Cryo-EM structure of 27a bound to E. coli RNAP and rrnBP1 promoter complex | | Descriptor: | (2S,7R,7aR,13aP,16Z,18E,20S,21S,22R,23R,24R,25S,26R,27S,28E)-5,21,23-trihydroxy-27-methoxy-2,4,16,20,22,24,26-heptamethyl-10-[4-(2-methylpropyl)piperazin-1-yl]-12-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1,6,15-trioxo-1,2,7,7a-tetrahydro-6H-2,7-(epoxypentadeca[1,11,13]trienoimino)[1]benzofuro[4,5-a]phenoxazin-25-yl acetate, DNA (5'-D(P*CP*TP*CP*GP*TP*AP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2021-11-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Optimization of Benzoxazinorifamycins to Improve Mycobacterium tuberculosis RNA Polymerase Inhibition and Treatment of Tuberculosis.
Acs Infect Dis., 8, 2022
|
|
7ML0
 
 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
1VY5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the post-catalysis state of peptide bond formation containing dipeptydil-tRNA in the A site and deacylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VY4
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5DK5
 
 | | Crystal structure of CRN-4-MES complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell death-related nuclease 4, ISOPROPYL ALCOHOL, ... | | Authors: | Hsiao, Y.-Y, Yuan, H.S. | | Deposit date: | 2015-09-03 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Inhibitors for the DEDDh Family of Exonucleases and a Unique Inhibition Mechanism by Crystal Structure Analysis of CRN-4 Bound with 2-Morpholin-4-ylethanesulfonate (MES)
J.Med.Chem., 59, 2016
|
|
3ULJ
 
 | | Crystal structure of apo Lin28B cold shock domain | | Descriptor: | ACETATE ION, GLYCEROL, Lin28b, ... | | Authors: | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | Deposit date: | 2011-11-10 | | Release date: | 2012-08-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The Lin28 cold-shock domain remodels pre-let-7 microRNA.
Nucleic Acids Res., 40, 2012
|
|
6ZTN
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTL
 
 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTJ
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
8WU8
 
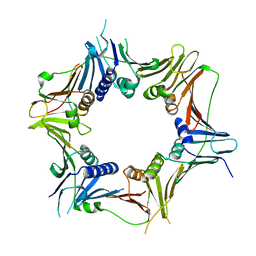 | | Crystal structure of the human RAD9-RAD1(F64A/M256A/F266A)-HUS1-RHINO(88-99) complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Hara, K, Nagata, K, Iida, N, Hashimoto, H. | | Deposit date: | 2023-10-20 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for intra- and intermolecular interactions on RAD9 subunit of 9-1-1 checkpoint clamp implies functional 9-1-1 regulation by RHINO.
J.Biol.Chem., 300, 2024
|
|
3LH0
 
 | |
6ZTO
 
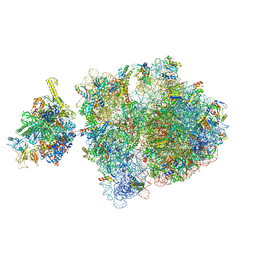 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZU1
 
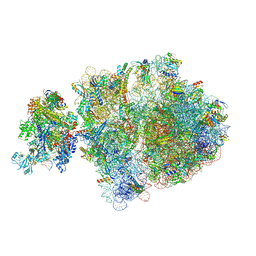 | | E. coli 70S-RNAP expressome complex in uncoupled state 2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
8SM3
 
 | | Structure of Bacillus cereus VD045 Gabija GajA-GajB Complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB, SULFATE ION | | Authors: | Antine, S.P, Mooney, S.E, Johnson, A.G, Kranzusch, P.J. | | Deposit date: | 2023-04-25 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
4UA2
 
 | | Crystal structure of dual function transcriptional regulator MerR from Bacillus megaterium MB1 | | Descriptor: | Regulatory protein | | Authors: | Lin, L.Y, Chang, C.C, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
4UA1
 
 | | Crystal structure of dual function transcriptional regulator MerR form Bacillus megaterium MB1 in complex with mercury (II) ion | | Descriptor: | MERCURY (II) ION, Regulatory protein | | Authors: | Chang, C.C, Lin, L.Y, Zou, X.W, Huang, C.C, Chan, N.L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the mercury(II)-mediated conformational switching of the dual-function transcriptional regulator MerR
Nucleic Acids Res., 43, 2015
|
|
6ZTM
 
 | | E. coli 70S-RNAP expressome complex in collided state without NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6QPJ
 
 | | Human CLOCK PAS-A domain | | Descriptor: | Circadian locomoter output cycles protein kaput | | Authors: | Kwon, H, Freeman, S.L, Moody, P.C.E, Raven, E.L, Basran, J. | | Deposit date: | 2019-02-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.315 Å) | | Cite: | Heme binding to human CLOCK affects interactions with the E-box.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
