5DT1
 
 | |
8A76
 
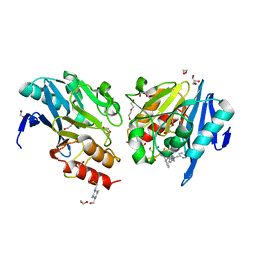 | | Metallo-beta-lactamase NDM-1 in complex with 1,2,4-Triazole-3-thione compound 26 | | Descriptor: | (2~{S})-2-[bis(pyridin-2-ylmethyl)amino]-5-(3-phenyl-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl)pentanoic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Vascon, F, Legru, A, Hernandez, J.F, Cendron, L. | | Deposit date: | 2022-06-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of 1,2,4-Triazole-3-thiones toward Broad-Spectrum Metallo-beta-lactamase Inhibitors Showing Potent Synergistic Activity on VIM- and NDM-1-Producing Clinical Isolates.
J.Med.Chem., 65, 2022
|
|
6S18
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with glycerol | | Descriptor: | Aromatic acid chemoreceptor, CHLORIDE ION, GLYCEROL | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S1A
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP | | Descriptor: | Aromatic acid chemoreceptor, SULFATE ION | | Authors: | Gavira, J.A, Matilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S3B
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with benzoate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aromatic acid chemoreceptor, ... | | Authors: | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | Deposit date: | 2019-06-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
7PZJ
 
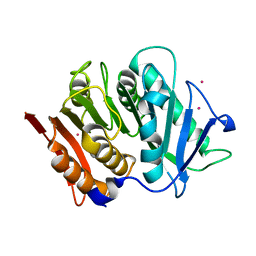 | | Structure of a bacteroidetal polyethylene terephthalate (PET) esterase | | Descriptor: | Lipase, POTASSIUM ION | | Authors: | Zang, H, Dierkes, R, Perez-Garcia, P, Weigert, S, Sternagel, S, Hallam, S.J, Applegate, V, Schumacher, J, Schott, T, Pleiss, J, Almeida, A, Hoecker, B, Smits, S.H, Schmitz, R.A, Chow, J, Streit, W.R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bacteroidetes Aequorivita sp. and Kaistella jeonii Produce Promiscuous Esterases With PET-Hydrolyzing Activity.
Front Microbiol, 12, 2021
|
|
6RRO
 
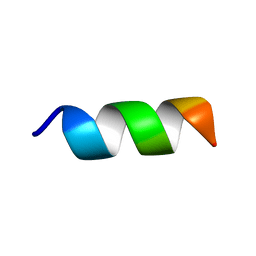 | |
6RRL
 
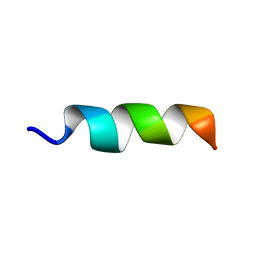 | |
6RSM
 
 | | Solution NMR structure of the peptide 12530 from medicinal leech Hirudo medicinalis in dodecylphosphocholine micelles | | Descriptor: | peptide 12530 | | Authors: | Talyzina, I.A, Nadezhdin, K.D, Grafskaia, E.N, Arseniev, A.S, Lazarev, V.N. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Medicinal leech antimicrobial peptides lacking toxicity represent a promising alternative strategy to combat antibiotic-resistant pathogens.
Eur.J.Med.Chem., 180, 2019
|
|
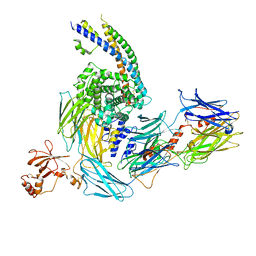
7ZGJ
 
 | |
7ZGK
 
 | |
7K0B
 
 | |
1U57
 
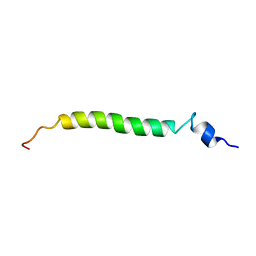 | | NMR structure of the (345-392)Gag sequence from HIV-1 | | Descriptor: | Gag polyprotein | | Authors: | Morellet, N, Druillennec, S, Lenoir, C, Bouaziz, S, Roques, B.P. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Helical structure determined by NMR of the HIV-1 (345-392)Gag sequence, surrounding p2: Implications for particle assembly and RNA packaging
Protein Sci., 14, 2005
|
|
7K5A
 
 | |
5AMX
 
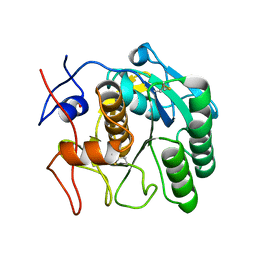 | | Crystal Structure of Proteinase K processed with the CrystalDirect automated mounting and cryo-cooling technology | | Descriptor: | PROTEINASE K, SULFATE ION | | Authors: | Zander, U, Hoffmann, G, Cornaciu, I, Cipriani, F, Marquez, J.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-04-13 | | Last modified: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Automated Harvesting and Processing of Protein Crystals Through Laser Photoablation.
Acta Crystallogr.,Sect.D, 72, 2016
|
|
3TC6
 
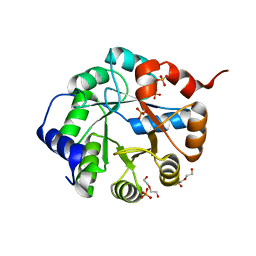 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
8DAX
 
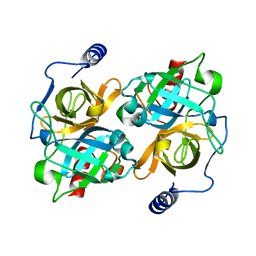 | | New insights into the P186 flip and oligomeric state of Staphylococcus aureus exfoliative toxin E: implications for the exfoliative mechanism | | Descriptor: | Exfoliative toxin E | | Authors: | Gismene, C, Nascimento, A.F.Z, Hernandez-Gonzalez, J.E, Santisteban, A.R.N, de Moraes, F.R, Arni, R.K, Mariutti, R.B. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Staphylococcus aureus Exfoliative Toxin E, Oligomeric State and Flip of P186: Implications for Its Action Mechanism.
Int J Mol Sci, 23, 2022
|
|
5EW1
 
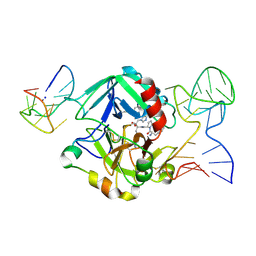 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT3, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
4QHI
 
 | | Crystal structure of Methanocaldococcus jannaschii selecase mutant R36W | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5EW2
 
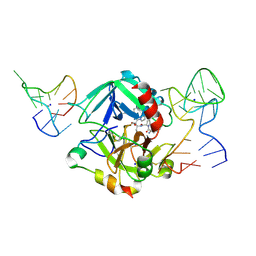 | | Human thrombin sandwiched between two DNA aptamers: HD22 and HD1-deltaT12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD1-deltaT12, ... | | Authors: | Pica, A, Russo Krauss, I, Parente, V, Sica, F. | | Deposit date: | 2015-11-20 | | Release date: | 2016-11-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers.
Nucleic Acids Res., 45, 2017
|
|
7PJM
 
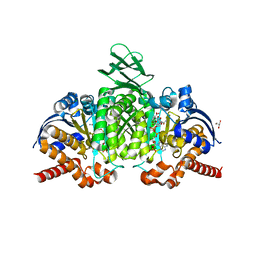 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
7PJN
 
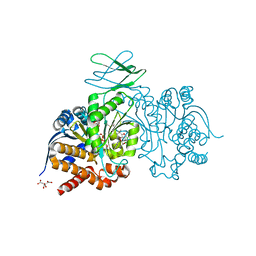 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
5MBD
 
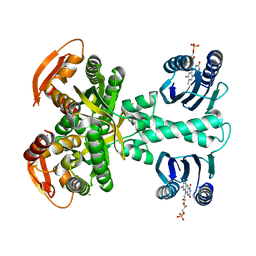 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MBJ
 
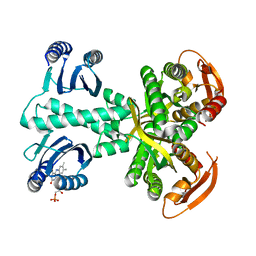 | | Structure of a bacterial light-regulated adenylyl cyclase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5APV
 
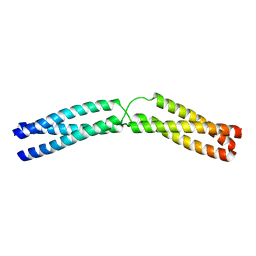 | |
