4P7H
 
 | |
7TQL
 
 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
8K2D
 
 | | Cryo-EM structure of the yeast 80S ribosome with tigecycline, eEF2, Stm1 and eIF5A | | Descriptor: | 18S rRNA, 23S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Beckmann, R, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
7KAI
 
 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8XSZ
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline, E-tRNA and P-tRNA | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2024-01-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
3THP
 
 | | Crystal structure and RNA binding properties of the RRM/AlkB domains in human ABH8, an enzyme catalyzing tRNA hypermodification, Northeast Structural Genomics Consortium Target HR5601B | | Descriptor: | 2-OXOGLUTARIC ACID, Alkylated DNA repair protein alkB homolog 8, MANGANESE (II) ION, ... | | Authors: | Pastore, C, Topalidou, I, Forouhar, F, Yan, A.C, Levy, M, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-19 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification.
J.Biol.Chem., 287, 2012
|
|
3J7A
 
 | | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine, small subunit | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Wong, W, Bai, X.C, Brown, A, Fernandez, I.S, Hanssen, E, Condron, M, Tan, Y.H, Baum, J, Scheres, S.H.W. | | Deposit date: | 2014-06-03 | | Release date: | 2014-07-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine.
Elife, 3, 2014
|
|
6D90
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-site tRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-27 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
7Z3N
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-1 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7MDZ
 
 | | 80S rabbit ribosome stalled with benzamide-CHX | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S21, ... | | Authors: | Koga, Y, Hoang, E.M, Park, Y, Keszei, A.F.A, Murray, J, Shao, S, Liau, B.B. | | Deposit date: | 2021-04-06 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Discovery of C13-Aminobenzoyl Cycloheximide Derivatives that Potently Inhibit Translation Elongation.
J.Am.Chem.Soc., 143, 2021
|
|
6T7I
 
 | | Structure of yeast 80S ribosome stalled on the CGA-CGA inhibitory codon combination. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Berninghausen, O, Becker, R, Beckmann, R. | | Deposit date: | 2019-10-22 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts.
Embo J., 39, 2020
|
|
6LQP
 
 | |
4U50
 
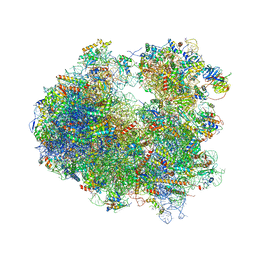 | | Crystal structure of Verrucarin bound to the yeast 80S ribosome | | Descriptor: | (4S,5R,10E,12Z,16R,16aS,17S,18R,19aR,23aR)-4-hydroxy-5,16a,21-trimethyl-4,5,6,7,16,16a,22,23-octahydro-3H,18H,19aH-spiro[16,18-methano[1,6,12]trioxacyclooctadecino[3,4-d]chromene-17,2'-oxirane]-3,9,14-trione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
4U4Y
 
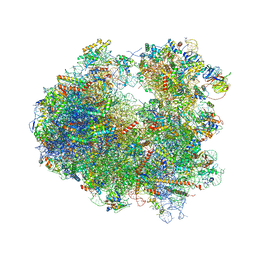 | | Crystal structure of Pactamycin bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2014-07-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
8JVI
 
 | | Structure of human TRPV4 with antagonist A2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
7UIY
 
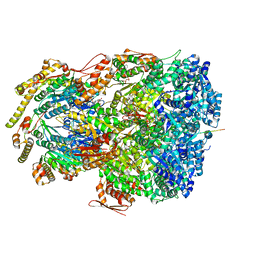 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8UT0
 
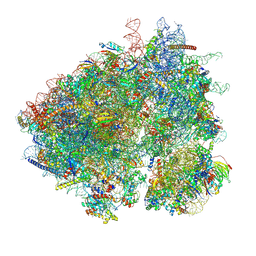 | | Eukaryotic 80S ribosome with Reh1, eIF5A and A/P site tRNA | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Yelland, J.N, Taylor, D.W, Johnson, A.W. | | Deposit date: | 2023-10-30 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The Ribosome Assembly Factor Reh1 is Released from the Polypeptide Exit Tunnel in the Pioneer Round of Translation
To Be Published
|
|
8IMY
 
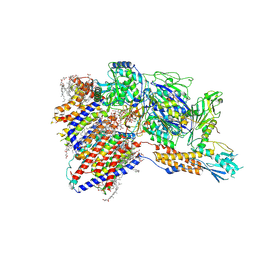 | | Cryo-EM structure of GPI-T (inactive mutant) with GPI and proULBP2, a proprotein substrate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, T, Xu, Y, Qu, Q, Li, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of liganded glycosylphosphatidylinositol transamidase illuminate GPI-AP biogenesis.
Nat Commun, 14, 2023
|
|
7XM9
 
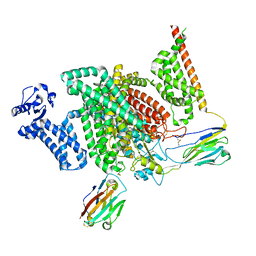 | | Cryo-EM structure of human NaV1.7/beta1/beta2-XEN907 | | Descriptor: | (7~{R})-1'-pentylspiro[6~{H}-furo[3,2-f][1,3]benzodioxole-7,3'-indole]-2'-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | zhang, J.T, Jiang, D.H. | | Deposit date: | 2022-04-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis for Na V 1.7 inhibition by pore blockers.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RR5
 
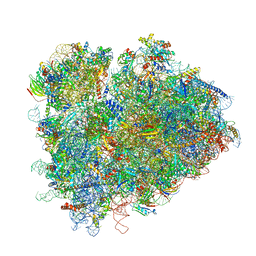 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
8G4I
 
 | | 40S ribosomal subunit of the 80S Giardia intestinalis assemblage A ribosome with Emetine bound in V1 conformation | | Descriptor: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S25, ... | | Authors: | Eiler, D.R, Wimberly, B.T, Bilodeau, D.Y, Rissland, O.S, Kieft, J.S. | | Deposit date: | 2023-02-09 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The Giardia lamblia ribosome structure reveals divergence in several biological pathways and the mode of emetine function.
Structure, 32, 2024
|
|
6UZ0
 
 | | Cardiac sodium channel with flecainide | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
7UIZ
 
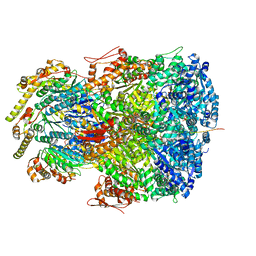 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
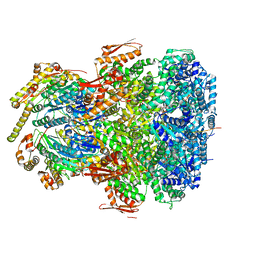 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5DC3
 
 | | Complex of yeast 80S ribosome with non-modified eIF5A | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Melnikov, S, Mailliot, J, Shin, B.-S, Rigger, L, Yusupova, G, Micura, R, Dever, T.E, Yusupov, M. | | Deposit date: | 2015-08-23 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structure of Hypusine-Containing Translation Factor eIF5A Bound to a Rotated Eukaryotic Ribosome.
J.Mol.Biol., 428, 2016
|
|
