6NCX
 
 | |
5FNS
 
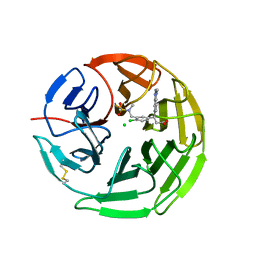 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5D60
 
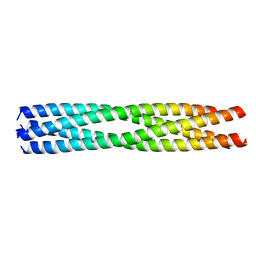 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form III | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7WGL
 
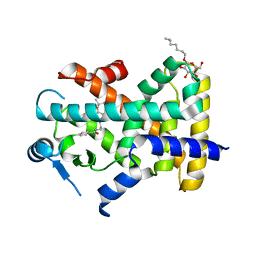 | | X-ray structure of human PPAR delta ligand binding domain-bezafibrate co-crystals obtained by co-crystallization | | Descriptor: | 2-[P-[2-P-CHLOROBENZAMIDO)ETHYL]PHENOXY]-2-METHYLPROPIONIC ACID, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Kamata, S, Honda, A, Machida, Y, Uchii, K, Shiiyama, Y, Masuda, R, Oyama, T, Ishii, I. | | Deposit date: | 2021-12-28 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Functional and Structural Insights into Human PPAR alpha / delta / gamma Subtype Selectivity of Bezafibrate, Fenofibric Acid, and Pemafibrate.
Int J Mol Sci, 23, 2022
|
|
5FMR
 
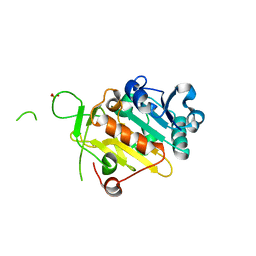 | | crIFT52 N-terminal domain | | Descriptor: | INTRAFLAGELLAR TRANSPORT PROTEIN COMPONENT IFT52, SULFATE ION | | Authors: | Mourao, A, Vetter, M, Lorentzen, E. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intraflagellar Transport Proteins 172, 80, 57, 54, 38, and 20 Form a Stable Tubulin-Binding Ift-B2 Complex.
Embo J., 35, 2016
|
|
5D69
 
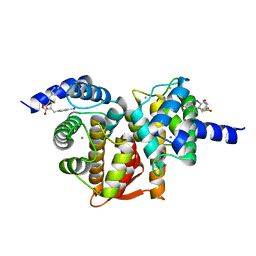 | | Human calpain PEF(S) with (2Z,2Z')-2,2'-disulfanediylbis(3-(6-iodoindol-3-yl)acrylic acid) bound | | Descriptor: | (2E,2'Z)-2,2'-disulfanediylbis[3-(4-iodophenyl)prop-2-enoic acid], CALCIUM ION, Calpain small subunit 1, ... | | Authors: | Adams, S.E, Robinson, E.J, Rizkallah, P.J, Miller, D.J, Hallett, M.B, Allemann, R.K. | | Deposit date: | 2015-08-11 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Conformationally restricted calpain inhibitors.
Chem Sci, 6, 2015
|
|
7PXO
 
 | | Structure of the Diels Alderase enzyme AbyU, from Micromonospora maris, co-crystallised with a non transformable substrate analogue | | Descriptor: | (2~{S},4~{S})-1-(4-methoxy-5-methyl-2-oxidanylidene-3~{H}-furan-3-yl)-2,4-dimethyl-dodecane-1,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Back, C.R, Race, P.R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Delineation of the Complete Reaction Cycle of a Natural Diels-Alderase
To Be Published
|
|
8VVM
 
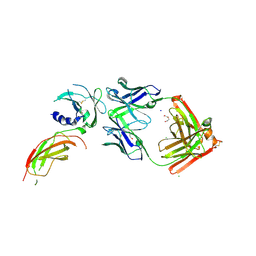 | | Structure of FabS1CE1-EPR1-1 in complex with the erythropoietin receptor | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Pavlenco, A, Ploder, L, Luu, G, Blazer, L, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antigen-binding fragments with improved crystal lattice packing and enhanced conformational flexibility at the elbow region as crystallization chaperones.
Protein Sci., 33, 2024
|
|
7O4T
 
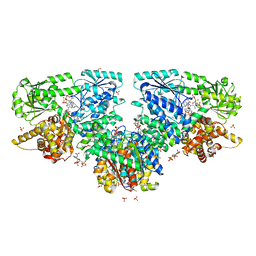 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and possible additional binding site (CoA(ECH/HAD)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7ZEA
 
 | |
7O1M
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7ZP2
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with BDA-04 | | Descriptor: | Aspartate carbamoyltransferase, SODIUM ION, SULFATE ION, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7QNP
 
 | | Designed Armadillo repeat protein N(A4)M4C(AII) co-crystallized with hen egg white lysozyme | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Designed Armadillo Repeat Protein N(A4)M4C(AII), ... | | Authors: | Michel, E, Mittl, P.R.E, Plueckthun, A. | | Deposit date: | 2021-12-21 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.586 Å) | | Cite: | Improved Repeat Protein Stability by Combined Consensus and Computational Protein Design.
Biochemistry, 62, 2023
|
|
8SLO
 
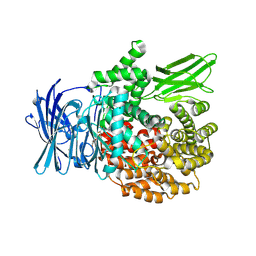 | | Plasmodium falciparum M1 aminopeptidase bound to selective inhibitor MIPS2673 | | Descriptor: | 2-hydroxy-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-2-methylpropanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | McGowan, S.M, Drinkwater, N. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.
Elife, 13, 2024
|
|
8PJ0
 
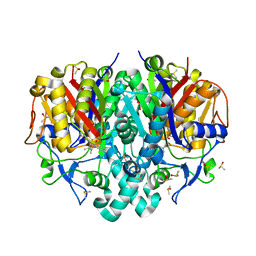 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-(1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazol-4-yl)-3-methylbutanamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Georgiou, C, Brenk, R, Espeland, L.O. | | Deposit date: | 2023-06-22 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, quality and validation of the EU-OPENSCREEN fragment library poised to a high-throughput screening collection.
Rsc Med Chem, 15, 2024
|
|
5VI4
 
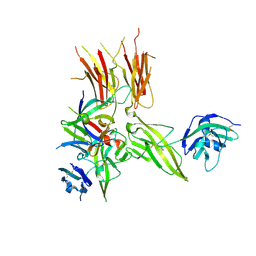 | | IL-33/ST2/IL-1RAcP ternary complex structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein, Interleukin-1 receptor-like 1, ... | | Authors: | Guenther, S, Sundberg, E.J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.792 Å) | | Cite: | IL-1 Family Cytokines Use Distinct Molecular Mechanisms to Signal through Their Shared Co-receptor.
Immunity, 47, 2017
|
|
7ZST
 
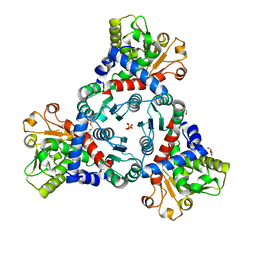 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with FLA-01 | | Descriptor: | 2-azanyl-~{N}-(2-methoxyethyl)-5-phenyl-thiophene-3-carboxamide, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-05-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
6NEO
 
 | |
8SM8
 
 | |
5FO8
 
 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
8SJ1
 
 | |
7ZH9
 
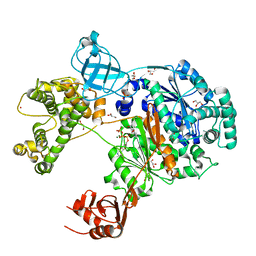 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
5VH0
 
 | | RHCC in complex with pyrene | | Descriptor: | 1,2-ETHANEDIOL, Tetrabrachion, pyrene | | Authors: | McDougall, M, Francisco, O, Stetefeld, J. | | Deposit date: | 2017-04-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Proteinaceous Nano container Encapsulate Polycyclic Aromatic Hydrocarbons.
Sci Rep, 9, 2019
|
|
7ZK1
 
 | |
7O6L
 
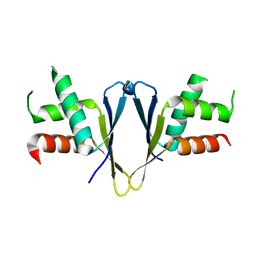 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
