3OZF
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with hypoxanthine | | Descriptor: | HYPOXANTHINE, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Ho, M, Hazleton, K.Z, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Acyclic Immucillin Phosphonates: Second-Generation Inhibitors of Plasmodium falciparum Hypoxanthine- Guanine-Xanthine Phosphoribosyltransferase.
Chem.Biol., 19, 2012
|
|
3P0J
 
 | |
3OSX
 
 | |
3OTC
 
 | |
3P2H
 
 | |
3OW6
 
 | | Crystal Structure of HSP90 with N-Aryl-benzimidazolone I | | Descriptor: | 1-(2,4-dihydroxyphenyl)-1,3-dihydro-2H-benzimidazol-2-one, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OWS
 
 | |
3OX1
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-{2-[7-(methylsulfamoyl)naphthalen-1-yl]ethyl}acetamide, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A, Mesecar, A.D. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3P0E
 
 | | Structure of hUPP2 in an active conformation with bound 5-benzylacyclouridine | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, Uridine phosphorylase 2 | | Authors: | Roosild, T.P, Castronovo, S, Villoso, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.
J.Struct.Biol., 176, 2011
|
|
3OYL
 
 | | Crystal structure of the PFV S217H mutant intasome bound to magnesium and the INSTI MK2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, AMMONIUM ION, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-23 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3P1T
 
 | |
3OZ7
 
 | |
3P31
 
 | |
3P0F
 
 | | Structure of hUPP2 in an inactive conformation with bound 5-benzylacyclouridine | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-BENZYLPYRIMIDINE-2,4(1H,3H)-DIONE, COBALT (II) ION, MAGNESIUM ION, ... | | Authors: | Roosild, T.P, Castronovo, S, Villoso, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A novel structural mechanism for redox regulation of uridine phosphorylase 2 activity.
J.Struct.Biol., 176, 2011
|
|
3P1U
 
 | |
3P3D
 
 | | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii | | Descriptor: | Nucleoporin 53 | | Authors: | Sampathkumar, P, Shawn, C, Bain, K, Gilmore, J, Gheyi, T, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-04 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the Nup53 RRM domain from Pichia guilliermondii
To be Published
|
|
3P7T
 
 | |
3P3P
 
 | |
3P4J
 
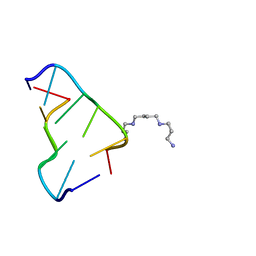 | | Ultra-high resolution structure of d(CGCGCG)2 Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE | | Authors: | Brzezinski, K, Brzuszkiewicz, A, Dauter, M, Kubicki, M, Jaskolski, M, Dauter, Z. | | Deposit date: | 2010-10-06 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.55 Å) | | Cite: | High regularity of Z-DNA revealed by ultra high-resolution crystal structure at 0.55 A.
Nucleic Acids Res., 39, 2011
|
|
3P4O
 
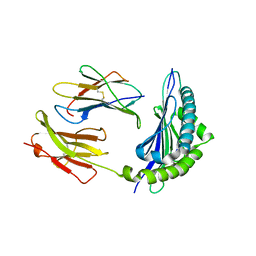 | | Crystal Structure of H2-Kb in complex with the mutant NP205-LCMV-V3A epitope YTAKYPNL, an 8-mer modified peptide from the LCMV | | Descriptor: | ACETYL GROUP, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Guillonneau, C, Rossjohn, J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of H2-Kb in complex with the mutant NP205-LCMV-V3A epitope YTAKYPNL, an 8-mer modified peptide from the LCMV
To be Published
|
|
3P54
 
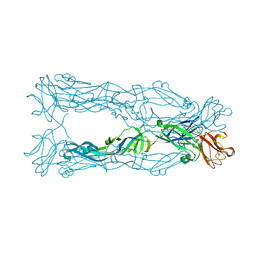 | | Crystal Structure of the Japanese Encephalitis Virus Envelope Protein, strain SA-14-14-2. | | Descriptor: | envelope glycoprotein | | Authors: | Luca, V.C, Nelson, C.A, AbiMansour, J.P, Diamond, M.S, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of the Japanese encephalitis virus envelope protein.
J.Virol., 86, 2012
|
|
3OW5
 
 | |
3OWH
 
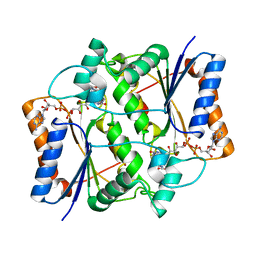 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Mesecar, A.D. | | Deposit date: | 2010-09-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
3P6W
 
 | |
3OWR
 
 | |
