5FFT
 
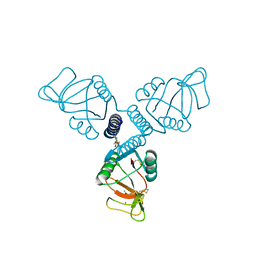 | | Crystal Structure of Surfactant Protein-A Y221A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
4O66
 
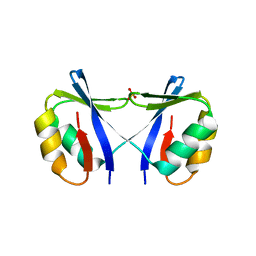 | | Crystal Structure of SMARCAL1 HARP substrate recognition domain | | Descriptor: | SODIUM ION, SULFATE ION, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 | | Authors: | Mason, A.C, Eichman, B.F. | | Deposit date: | 2013-12-20 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structure-specific nucleic acid-binding domain conserved among DNA repair proteins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6RGK
 
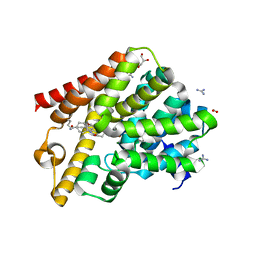 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-055 | | Descriptor: | 3-[5-[(4~{a}~{R},8~{a}~{S})-3-cycloheptyl-4-oxidanylidene-4~{a},5,6,7,8,8~{a}-hexahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-butyl-prop-2-ynamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-16 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
7UC5
 
 | |
5LXM
 
 | |
2WPW
 
 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (without AcCoA) | | Descriptor: | ACETYL COENZYME *A, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
6RB6
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-053 | | Descriptor: | 1,2-ETHANEDIOL, 3-[5-[(4~{a}~{R},8~{a}~{S})-3-cycloheptyl-4-oxidanylidene-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(furan-2-ylmethyl)prop-2-ynamide, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
6RCW
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-053 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-[5-[(4~{a}~{R},8~{a}~{S})-3-cycloheptyl-4-oxidanylidene-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(furan-2-ylmethyl)prop-2-ynamide, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-11 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
6SEF
 
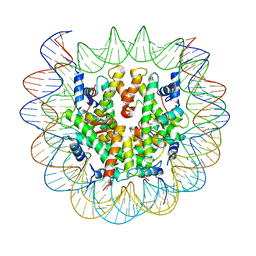 | | Class2C : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
5TC4
 
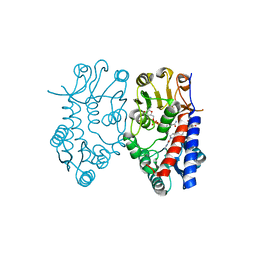 | | Crystal structure of human mitochondrial methylenetetrahydrofolate dehydrogenase-cyclohydrolase (MTHFD2) in complex with LY345899 and cofactors | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Jemth, A.-S, Gustafsson Sheppard, N, Farnegardh, K, Loseva, O, Wiita, E, Bonagas, N, Dahllund, L, Llona-Minguez, S, Haggblad, M, Henriksson, M, Andersson, Y, Homan, E, Helleday, T, Stenmark, P. | | Deposit date: | 2016-09-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of the Emerging Cancer Target MTHFD2 in Complex with a Substrate-Based Inhibitor.
Cancer Res., 77, 2017
|
|
821P
 
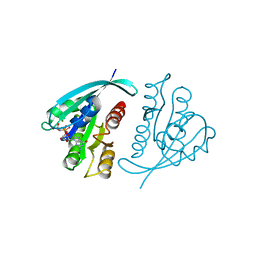 | | THREE-DIMENSIONAL STRUCTURES AND PROPERTIES OF A TRANSFORMING AND A NONTRANSFORMING GLYCINE-12 MUTANT OF P21H-RAS | | Descriptor: | C-H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Scheidig, A.J, Krengel, U, Pai, E.F, Kabsch, W, Wittinghofer, A, Goody, R.S. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras.
Biochemistry, 32, 1993
|
|
6QX8
 
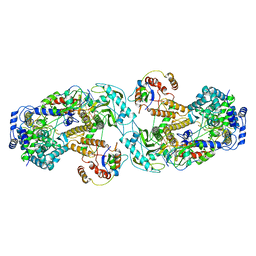 | | Influenza A virus (A/NT/60/1968) polymerase dimer of heterotrimer in complex with 5' cRNA promoter | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*CP*AP*AP*AP*AP*GP*CP*AP*GP*A)-3'), ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
1B20
 
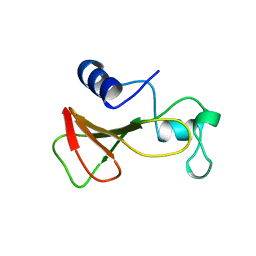 | | DELETION OF A BURIED SALT-BRIDGE IN BARNASE | | Descriptor: | PROTEIN (BARNASE), ZINC ION | | Authors: | Vaughan, C.K, Harryson, P, Buckle, A.M, Oliveberg, M, Fersht, A.R. | | Deposit date: | 1998-12-03 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structural double-mutant cycle: estimating the strength of a buried salt bridge in barnase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5C00
 
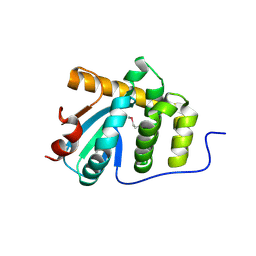 | | MdbA protein, a thiol-disulfide oxidoreductase from Corynebacterium diphtheriae | | Descriptor: | MdbA protein | | Authors: | OSIPIUK, J, REARDON-ROBINSON, M.E, TON-THAT, H, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-06-11 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A thiol-disulfide oxidoreductase of the Gram-positive pathogen Corynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence.
Mol.Microbiol., 98, 2015
|
|
3QRQ
 
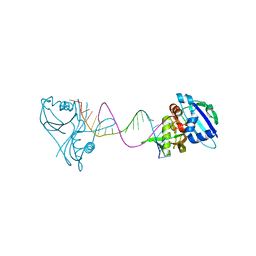 | | Structure of Thermus Thermophilus Cse3 bound to an RNA representing a pre-cleavage complex | | Descriptor: | Putative uncharacterized protein TTHB192, RNA (5'-R(*GP*UP*CP*CP*CP*CP*AP*CP*GP*CP*GP*UP*GP*UP*GP*GP*GP*GP*A)-3') | | Authors: | Schellenberg, M.J, Gesner, E.G, Garside, E.L, MacMillan, A.M. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Recognition and maturation of effector RNAs in a CRISPR interference pathway.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1ARM
 
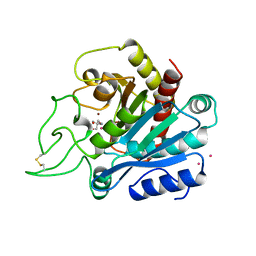 | | CARBOXYPEPTIDASE A WITH ZN REPLACED BY HG | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, HG-CARBOXYPEPTIDASE A=ALPHA= (COX), ... | | Authors: | Greenblatt, H.M, Feinberg, H, Tucker, P.A, Shoham, G. | | Deposit date: | 1994-11-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Carboxypeptidase A: native, zinc-removed and mercury-replaced forms.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
7ZHH
 
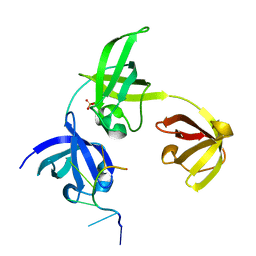 | | Complex structure of drosophila Unr CSD789 and a poly(A) RNA sequence | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*A)-3'), SULFATE ION, Upstream of N-ras, ... | | Authors: | Hollmann, N.M, Jagtap, P.K.A, Hennig, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Nucleic Acids Res., 51, 2023
|
|
8YVO
 
 | | Crystal structure of the C. difficile toxin A CROPs domain fragment 2639-2707 bound to C4.2 nanobody | | Descriptor: | 1,2-ETHANEDIOL, C4.2 nanobody, Toxin A | | Authors: | Sluchanko, N.N, Varfolomeeva, L.A, Shcheblyakov, D.V, Belyi, Y.F, Logunov, D.Y, Gintsburg, A.L, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into recognition of Clostridioides difficile toxin A by novel neutralizing nanobodies targeting QTIN-like motifs within its receptor-binding domain.
Int.J.Biol.Macromol., 283, 2024
|
|
2WPX
 
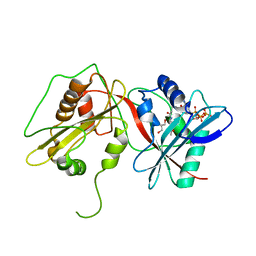 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
6EQG
 
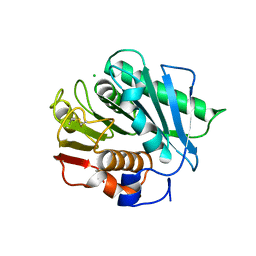 | | Crystal structure of a polyethylene terephthalate degrading hydrolase from Ideonella sakaiensis in spacegroup P21 | | Descriptor: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Austin, H.P, Allen, M.D, Johnson, C.W, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2017-10-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization and engineering of a plastic-degrading aromatic polyesterase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
1BDD
 
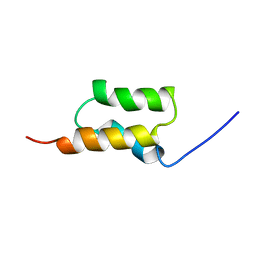 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
1BH4
 
 | |
5N1X
 
 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-ethyl-5-pyridin-3-yl-pyrazolo[1,5-a]pyrimidin-7-amine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
1BDC
 
 | | STAPHYLOCOCCUS AUREUS PROTEIN A, IMMUNOGLOBULIN-BINDING B DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCUS AUREUS PROTEIN A | | Authors: | Gouda, H, Torigoe, H, Saito, A, Sato, M, Arata, Y, Shimada, I. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures.
Biochemistry, 31, 1992
|
|
