2LR1
 
 | |
4C1Y
 
 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with b-methylfucoside | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, FUCOSE-SPECIFIC LECTIN FLEA, ... | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2013-08-14 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5VAU
 
 | |
5UYP
 
 | | 70S ribosome bound with near-cognate ternary complex base-paired to A site codon, open 30S (Structure II-nc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
4CA2
 
 | |
4CCX
 
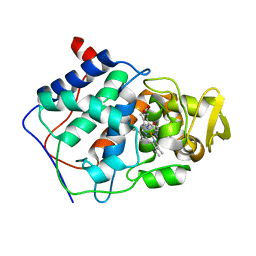 | | ALTERING SUBSTRATE SPECIFICITY AT THE HEME EDGE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wilcox, S.K, Jensen, G.M, Fitzgerald, M.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-03-17 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Altering substrate specificity at the heme edge of cytochrome c peroxidase.
Biochemistry, 35, 1996
|
|
6TIQ
 
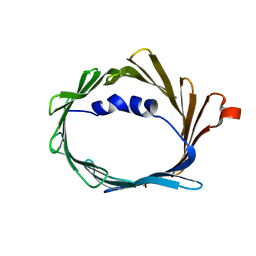 | |
4C39
 
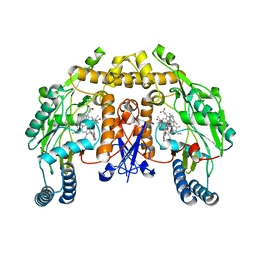 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-((((3S, 5R)-5-(((6-amino-4-methylpyridin-2-yl)methoxy) methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-((((3S, 5R)-5-(((6-amino-4-methylpyridin-2-yl)methoxy)methyl)pyrrolidin-3-yl)oxy)methyl)-4-methylpyridin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-08-22 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An Accessible Chiral Linker to Enhance Potency and Selectivity of Neuronal Nitric Oxide Synthase Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
6TF6
 
 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | CHLORIDE ION, Galectin-3, ~{N}-[[(2~{S},3~{S},4~{R},5~{S},6~{R})-4-[[5,6-bis(fluoranyl)-2-oxidanylidene-chromen-3-yl]methoxy]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]methyl]-4-fluoranyl-naphthalene-1-carboxamide | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
6TF9
 
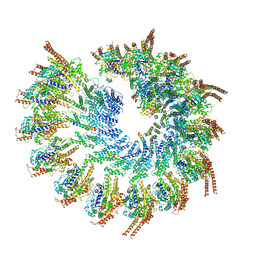 | | Structure of the vertebrate gamma-Tubulin Ring Complex | | Descriptor: | Actin, cytoplasmic 1, Belt helices 1,2,3,4, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2019-11-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Insights into the assembly and activation of the microtubule nucleator gamma-TuRC.
Nature, 578, 2020
|
|
2LAG
 
 | | Structure of the 44 kDa complex of interferon-alpha2 with the extracellular part of IFNAR2 obtained by 2D-double difference NOESY | | Descriptor: | Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Nudelman, I, Akabayov, S.R, Scherf, T, Anglister, J. | | Deposit date: | 2011-03-13 | | Release date: | 2011-08-17 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Observation of Intermolecular Interactions in Large Protein Complexes by 2D-Double Difference Nuclear Overhauser Enhancement Spectroscopy: Application to the 44 kDa Interferon-Receptor Complex.
J.Am.Chem.Soc., 133, 2011
|
|
5UYK
 
 | | 70S ribosome bound with cognate ternary complex not base-paired to A site codon (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ensemble cryo-EM elucidates the mechanism of translation fidelity
Nature, 546, 2017
|
|
6T2L
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, GLYCEROL, ... | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
5V59
 
 | |
6T30
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, GLYCEROL, ... | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
2OA6
 
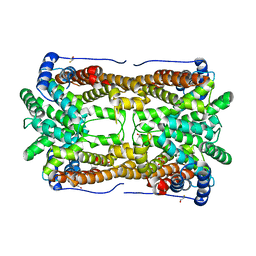 | | Aristolochene synthase from Aspergillus terreus complexed with pyrophosphate | | Descriptor: | Aristolochene synthase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Shishova, E.Y, Di Costanzo, L, Cane, D.E, Christianson, D.W. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate
Biochemistry, 46, 2007
|
|
2OCC
 
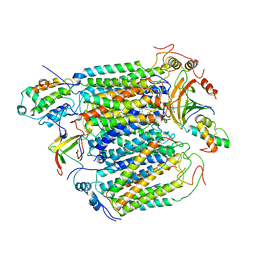 | |
2OBC
 
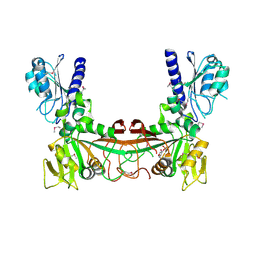 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
5UJE
 
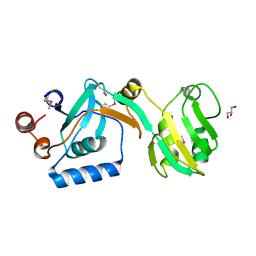 | |
4DNE
 
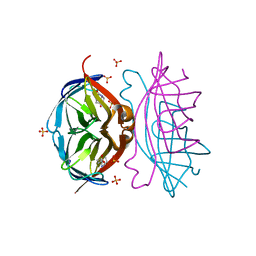 | | Crystal structure of a triple-mutant of streptavidin in complex with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, SULFATE ION, Streptavidin | | Authors: | Panwar, P, Deniaud, A, Pebay-Peyroula, E. | | Deposit date: | 2012-02-08 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Contamination from an affinity column: an encounter with a new villain in the world of membrane-protein crystallization.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5UNR
 
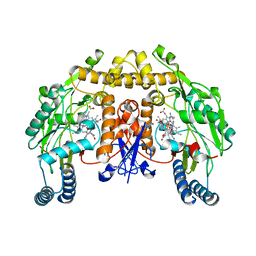 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 3-[(2-aminoquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 3-[(2-aminoquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UO5
 
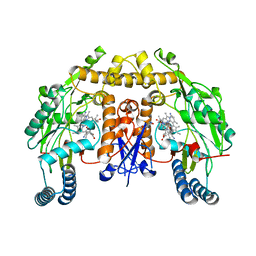 | | Structure of human neuronal nitric oxide synthase heme domain in complex with (RS)-3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-(2-(methylamino)propyl)benzonitrile | | Descriptor: | 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2R)-2-(methylamino)propyl]benzonitrile, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(2S)-2-(methylamino)propyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
2OIE
 
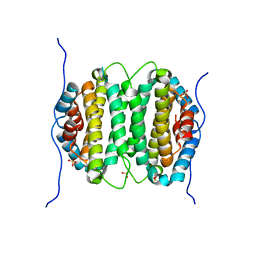 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
5UUK
 
 | |
4DBW
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 (AKR1C3) in complex with NADP+ and 2'-desmethyl-indomethacin | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [1-(4-chlorobenzoyl)-5-methoxy-1H-indol-3-yl]acetic acid | | Authors: | Chen, M, Christianson, D.W, Marnett, L.J, Penning, T.M. | | Deposit date: | 2012-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Development of potent and selective indomethacin analogues for the inhibition of AKR1C3 (Type 5 17 beta-hydroxysteroid dehydrogenase/prostaglandin F synthase) in castrate-resistant prostate cancer.
J.Med.Chem., 56, 2013
|
|
