3L81
 
 | | Crystal structure of adaptor protein complex 4 (AP-4) mu4 subunit C-terminal domain, in complex with a sorting peptide from the amyloid precursor protein (APP) | | Descriptor: | AP-4 complex subunit mu-1, Amyloid beta A4 protein, GLYCEROL | | Authors: | Mardones, G.A, Rojas, A.L, Burgos, P.V, Dasilva, L.L.P, Prabhu, Y, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2009-12-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sorting of the Alzheimer's disease amyloid precursor protein mediated by the AP-4 complex.
Dev.Cell, 18, 2010
|
|
5EAR
 
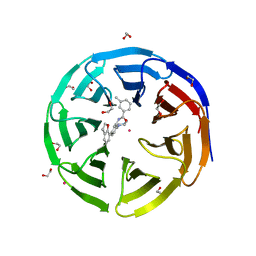 | | Crystal structure of human WDR5 in complex with compound 9d | | Descriptor: | 1,2-ETHANEDIOL, N-[5-(2,3-dihydro-1-benzofuran-7-yl)-2-(4-methylpiperazin-1-yl)phenyl]-3-methylbenzamide, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
8PD6
 
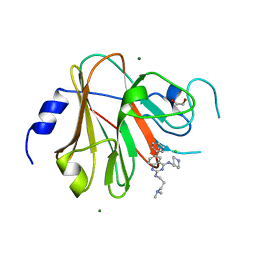 | | Crystal structure of the TRIM58 PRY-SPRY domain in complex with TRIM-473 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase TRIM58, ... | | Authors: | Renatus, M, Hoegenauer, K, Schroeder, M. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of Ligands for TRIM58, a Novel Tissue-Selective E3 Ligase.
Acs Med.Chem.Lett., 14, 2023
|
|
8PD4
 
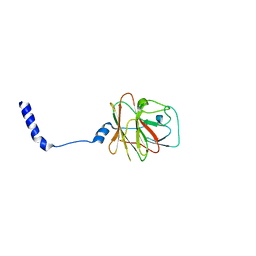 | |
5HTZ
 
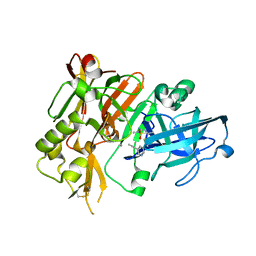 | | BACE1 in complex with (S)-5-(3-chloro-5-(5-(prop-1-yn-1-yl)pyridin-3-yl)thiophen-2-yl)-2,5-dimethyl-1,2,4-thiadiazinan-3-iminium 1,1-dioxide | | Descriptor: | (3E,5S)-5-{3-chloro-5-[5-(prop-1-yn-1-yl)pyridin-3-yl]thiophen-2-yl}-2,5-dimethyl-1,2,4-thiadiazinan-3-imine 1,1-dioxide, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
7SHU
 
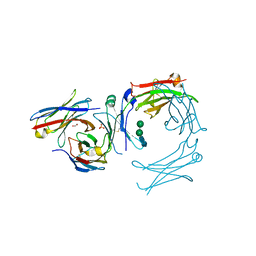 | | IgE-Fc in complex with omalizumab variant C02 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant epsilon, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SHY
 
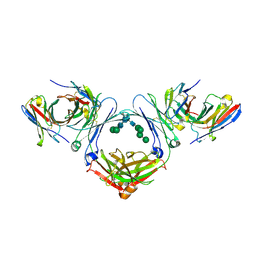 | | IgE-Fc in complex with omalizumab scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SI0
 
 | | IgE-Fc in complex with 813 | | Descriptor: | 813 Variable fragment Heavy chain, 813 Variable fragment Light chain, IgE Fc, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
7SHZ
 
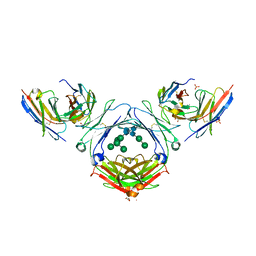 | | IgE-Fc in complex with HAE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Pennington, L.F, Jardetzky, T.J, Kleinboelting, S. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Directed evolution of and structural insights into antibody-mediated disruption of a stable receptor-ligand complex.
Nat Commun, 12, 2021
|
|
3LQH
 
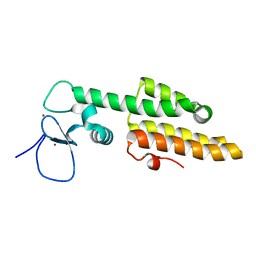 | | Crystal structure of MLL1 PHD3-Bromo in the free form | | Descriptor: | Histone-lysine N-methyltransferase MLL, ZINC ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-09 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
5EAM
 
 | | Crystal structure of human WDR5 in complex with compound 9o | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, ... | | Authors: | DONG, A, DOMBROVSKI, L, SMIL, D, GETLIK, M, BOLSHAN, Y, WALKER, J.R, SENISTERRA, G, PODA, G, AL-AWAR, R, SCHAPIRA, M, VEDADI, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-10-16 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of a Small Molecule Antagonist of the Interaction Between WD Repeat-Containing Protein 5 (WDR5) and Mixed-Lineage Leukemia 1 (MLL1).
J. Med. Chem., 59, 2016
|
|
5L3A
 
 | | Fragment-based discovery of 6-arylindazole JAK inhibitors | | Descriptor: | Tyrosine-protein kinase JAK2, ~{N}-(1~{H}-indazol-4-yl)methanesulfonamide | | Authors: | Soerensen, M.D, Dack, K.N, Greve, D.R, Ritzen, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Fragment-Based Discovery of 6-Arylindazole JAK Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5EML
 
 | |
5EMJ
 
 | | Crystal structure of PRMT5:MEP50 with Compound 8 and sinefungin | | Descriptor: | (2~{S})-1-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-3-[[4-(3-methylbenzimidazol-5-yl)pyridin-2-yl]amino]propan-2-ol, GLYCEROL, Methylosome protein 50, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structure and Property Guided Design in the Identification of PRMT5 Tool Compound EPZ015666.
ACS Med Chem Lett, 7, 2016
|
|
8A3Y
 
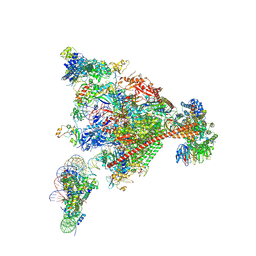 | | Structure of mammalian Pol II-DSIF-SPT6-PAF1-TFIIS-hexasome elongation complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Farnung, L, Ochmann, M, Garg, G, Vos, S.M, Cramer, P. | | Deposit date: | 2022-06-09 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a backtracked hexasomal intermediate of nucleosome transcription.
Mol.Cell, 82, 2022
|
|
5IST
 
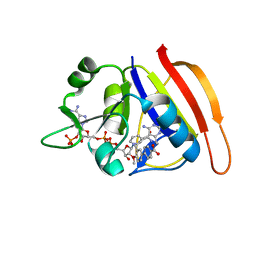 | | Staphylococcus aureus Dihydrofolate Reductase complexed with beta-NADPH, cyclic alpha-NADPH anomer and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.723 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
5ISQ
 
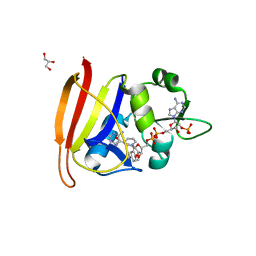 | | Staphylococcus aureus H30N, F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
5ISP
 
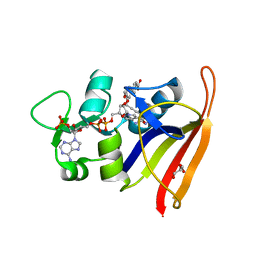 | | Staphylococcus aureus F98Y Dihydrofolate Reductase mutant complexed with beta-NADPH and 3'-(3-(2,4-diamino-6-ethylpyrimidin-5-yl)prop-2-yn-1-yl)-4'-methoxy-[1,1'-biphenyl]-4-carboxylic acid (UCP1106) | | Descriptor: | 4-[3-[3-[2,4-bis(azanyl)-6-ethyl-pyrimidin-5-yl]prop-2-ynyl]-4-methoxy-phenyl]benzoic acid, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Anderson, A.C, Reeve, S.M. | | Deposit date: | 2016-03-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Charged Propargyl-Linked Antifolates Reveal Mechanisms of Antifolate Resistance and Inhibit Trimethoprim-Resistant MRSA Strains Possessing Clinically Relevant Mutations.
J. Med. Chem., 59, 2016
|
|
6T1M
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with benzimidazole-amide derivative 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-cyano-~{N}-[2-(piperidin-1-ylmethyl)-1~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Chaikuad, A, Heidenreich, D, Moustakim, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3ZO2
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(2,9-Diazaspiro[5.5]undecan-9-yl)-9H-purine, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
5EOL
 
 | |
6T1J
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3ZO4
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(4-PHENYL-1,9-DIAZASPIRO[5.5]UNDECAN-9-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
6T1L
 
 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 3 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(diethylaminomethyl)phenyl]methyl]-4-pyrimidin-2-yl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
3ZO3
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | 6-(2,9-DIAZASPIRO[5.5]UNDECAN-2-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
