7AJU
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
5I59
 
 | | Glutamate- and glycine-bound GluN1/GluN2A agonist binding domains with MPX 007 | | Descriptor: | 5-({[(3,4-difluorophenyl)sulfonyl]amino}methyl)-6-methyl-N-[(2-methyl-4H-1lambda~4~,3-thiazol-5-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Mou, T.-C, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2016-02-14 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Negative Allosteric Modulation of GluN2A-Containing NMDA Receptors.
Neuron, 91, 2016
|
|
4X8J
 
 | | Crystal Structure of murine 12F4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | 12F4 FAB Heavy chain, 12F4 FAB Light chain, NONAETHYLENE GLYCOL, ... | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
8HTY
 
 | | Candida boidinii Formate Dehydrogenase Crystal Structure at 1.4 Angstrom Resolution | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Gul, M, Yuksel, B, Bulut, H, DeMirci, H. | | Deposit date: | 2022-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of wild-type and Val120Thr mutant Candida boidinii formate dehydrogenase by X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5SB5
 
 | | Tubulin-todalam-9-complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2021-07-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7AHC
 
 | | OpuA apo inward-facing | | Descriptor: | ABC transporter permease subunit, ABC-type proline/glycine betaine transport system ATPase component | | Authors: | Sikkema, H.R, Rheinberger, J, Paulino, C, Poolman, B. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Gating by ionic strength and safety check by cyclic-di-AMP in the ABC transporter OpuA.
Sci Adv, 6, 2020
|
|
7K9H
 
 | | SARS-CoV-2 Spike in complex with neutralizing Fab 2B04 (one up, two down conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B04 heavy chain, ... | | Authors: | Errico, J.M, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SARS-CoV-2 neutralization by two murine antibodies targeting the RBD.
Cell Rep, 37, 2021
|
|
5IAB
 
 | | Caspase 3 V266D | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASK, Caspase-3 | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7AJT
 
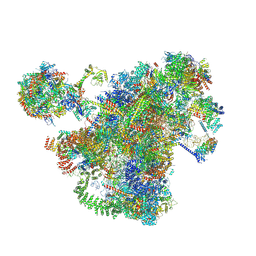 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
4WKR
 
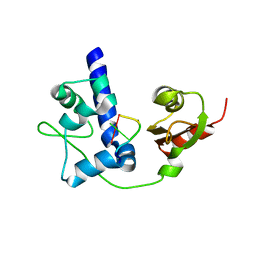 | | LaRP7 wrapping up the 3' hairpin of 7SK non-coding RNA (302-332) | | Descriptor: | 7SK GGHP4 (300-332), La-related protein 7 | | Authors: | Uchikawa, E, Natchiar, K.S, Han, X, Proux, F, Roblin, P, Zhang, E, Durand, A, Klaholz, B.P, Dock-Bregeon, A.-C. | | Deposit date: | 2014-10-03 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the mechanism of stabilization of the 7SK small nuclear RNA by LARP7.
Nucleic Acids Res., 43, 2015
|
|
6EK3
 
 | |
7AHJ
 
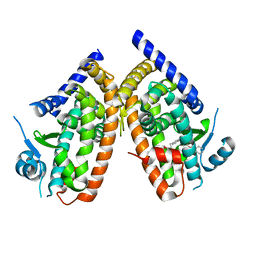 | | Crystal structure of PPARgamma V290M mutant ligand binding domain in complex with farglitazar | | Descriptor: | 2-(2-BENZOYL-PHENYLAMINO)-3-{4-[2-(5-METHYL-2-PHENYL-OXAZOL-4-YL)-ETHOXY]-PHENYL}-PROPIONIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Schoenmakers, E, Schwabe, B.T.W, Fairall, L, Chatterjee, K, Schwabe, J.W.R. | | Deposit date: | 2020-09-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PPARgamma V290M mutant ligand binding domain in complex with farglitazar
To Be Published
|
|
7AO0
 
 | | Crystal structure of CotB2 variant F107A in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
4WLH
 
 | | High resolution crystal structure of human kynurenine aminotransferase-I bound to PLP cofactor | | Descriptor: | Kynurenine--oxoglutarate transaminase 1 | | Authors: | Nadvi, N.A, Salam, N.K, Park, J, Akladios, F.N, Kapoor, V, Collyer, C.A, Gorrell, M.D, Church, W.B. | | Deposit date: | 2014-10-07 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | High resolution crystal structures of human kynurenine aminotransferase-I bound to PLP cofactor, and in complex with aminooxyacetate.
Protein Sci., 26, 2017
|
|
5AR2
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) | | Descriptor: | CALCIUM ION, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
7ZVX
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 2.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, ... | | Authors: | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | Deposit date: | 2022-05-17 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
8ENQ
 
 | | E. coli CsgA fibril (218-pixel box size) | | Descriptor: | Major curlin subunit | | Authors: | Bu, F, Liu, B. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insight into Escherichia coli CsgA amyloid fibril assembly.
Mbio, 15, 2024
|
|
6ECM
 
 | |
5KM9
 
 | |
1BDX
 
 | | E. COLI DNA HELICASE RUVA WITH BOUND DNA HOLLIDAY JUNCTION, ALPHA CARBONS AND PHOSPHATE ATOMS ONLY | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*GP*CP*AP*TP*AP*TP*GP*CP*AP*TP*GP*C)-3'), HOLLIDAY JUNCTION DNA HELICASE RUVA | | Authors: | Hargreaves, D, Rice, D.W, Sedelnikova, S.E, Artymiuk, P.J, Lloyd, R.G, Rafferty, J.B. | | Deposit date: | 1998-05-11 | | Release date: | 1999-11-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structure of E.coli RuvA with bound DNA Holliday junction at 6 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
6UNW
 
 | | Epoxide hydrolase from an endophytic Streptomyces | | Descriptor: | CACODYLATE ION, CHLORIDE ION, Soluble epoxide hydrolase | | Authors: | Wilson, C, dos Santos, J.C, Dias, M.V.B. | | Deposit date: | 2019-10-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | An epoxide hydrolase from endophytic Streptomyces shows unique structural features and wide biocatalytic activity.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5DZI
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3S)-3-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[(3S)-3-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5I2F
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) with bound sulfamide inhibitor Bio-AMS | | Descriptor: | 1,2-ETHANEDIOL, 5'-deoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Histidine triad nucleotide-binding protein 1 | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design, Synthesis, and Characterization of Sulfamide and Sulfamate Nucleotidomimetic Inhibitors of hHint1.
Acs Med.Chem.Lett., 7, 2016
|
|
1BA1
 
 | |
1B8C
 
 | | PARVALBUMIN | | Descriptor: | MAGNESIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
