7UW4
 
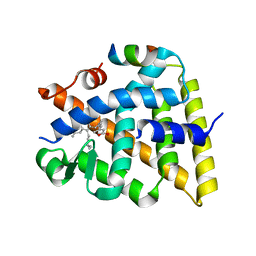 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB113 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-propylcyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
5DUQ
 
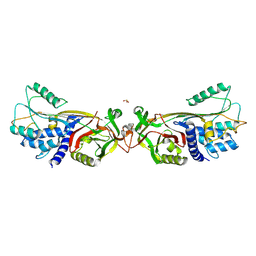 | | Active human c1-inhibitor in complex with dextran sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plasma protease C1 inhibitor, SULFITE ION, ... | | Authors: | Dijk, M, Holkers, J, Voskamp, P, Giannetti, B.M, Waterreus, W.J, van Veen, H.A, Pannu, N.S. | | Deposit date: | 2015-09-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | How Dextran Sulfate Affects C1-inhibitor Activity: A Model for Polysaccharide Potentiation.
Structure, 24, 2016
|
|
8T2O
 
 | |
8T2B
 
 | |
7N47
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
5EBB
 
 | | Structure of human sphingomyelinase phosphodiesterase like 3A (SMPDL3A) with Zn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, GLYCEROL, ... | | Authors: | Lim, S.M, Yeung, K, Tresaugues, L, Teo, H.L, Nordlund, P. | | Deposit date: | 2015-10-19 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and catalytic mechanism of human sphingomyelin phosphodiesterase like 3a - an acid sphingomyelinase homologue with a novel nucleotide hydrolase activity.
Febs J., 283, 2016
|
|
5VIC
 
 | | Crystal structure of anti-Zika antibody Z004 bound to DENV-1 Envelope protein DIII | | Descriptor: | Dengue 1 Envelope DIII domain, Fab heavy chain, Fab light chain | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
7MY0
 
 | | Sy-CrtE IPP structure | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
8PWN
 
 | | Structure of A2A adenosine receptor A2AR-StaR2-bRIL, solved at wavelength 2.75 A | | Descriptor: | Adenosine receptor A2a,Soluble cytochrome b562, CHOLESTEROL, OLEIC ACID, ... | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Romano, M, Moraes, I, Wagner, A. | | Deposit date: | 2023-07-20 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6NPE
 
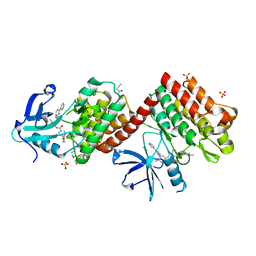 | | C-abl Kinase domain with the activator(cmpd6), 2-cyano-N-(4-(3,4-dichlorophenyl)thiazol-2-yl)acetamide | | Descriptor: | 2-cyano-~{N}-[4-(3,4-dichlorophenyl)-1,3-thiazol-2-yl]ethanamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, NONAETHYLENE GLYCOL, ... | | Authors: | campobasso, N. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Identification and Optimization of Novel Small c-Abl Kinase Activators Using Fragment and HTS Methodologies.
J. Med. Chem., 62, 2019
|
|
7ULY
 
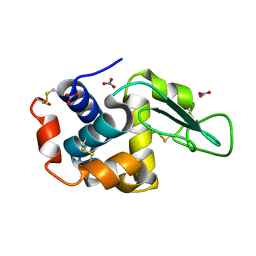 | | MicroED structure of triclinic lysozyme | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Gonen, T. | | Deposit date: | 2022-04-05 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.87 Å) | | Cite: | Hydrogens and hydrogen-bond networks in macromolecular MicroED data.
J Struct Biol X, 6, 2022
|
|
8PXK
 
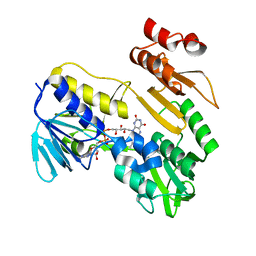 | | Structure of NADH-DEPENDENT FERREDOXIN REDUCTASE, BPHA4, solved at wavelength 5.76 A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin reductase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Senda, M, Matsugaki, N, Kawano, Y, Wagner, A. | | Deposit date: | 2023-07-23 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
7YPZ
 
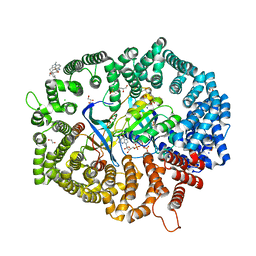 | | Zafirlukast in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Searching for Novel Noncovalent Nuclear Export Inhibitors through a Drug Repurposing Approach.
J.Med.Chem., 66, 2023
|
|
5HY2
 
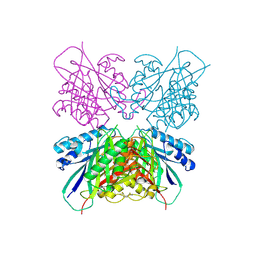 | | Structure-function analysis of functionally diverse members of the cyclic amide hydrolase family of Toblerone fold enzymes | | Descriptor: | Ring-opening amidohydrolase | | Authors: | Peat, T.S, Balotra, S, Wilding, M, Newman, J, Scott, C. | | Deposit date: | 2016-02-01 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-Resolution X-Ray Structures of Two Functionally Distinct Members of the Cyclic Amide Hydrolase Family of Toblerone Fold Enzymes.
Appl. Environ. Microbiol., 83, 2017
|
|
8VF0
 
 | |
8T2A
 
 | |
8PX1
 
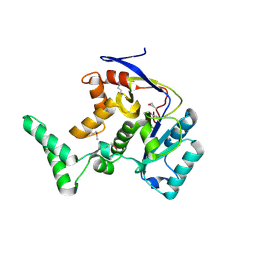 | | Structure of salmonella effector SseK3, solved at wavelength 2.75 A | | Descriptor: | Non-LEE encoded effector protein NleB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C.M, Esposito, D, Rittinger, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
6NQF
 
 | |
8PNW
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
7PBC
 
 | | Crystal structure of engineered TCR (796) complexed to HLA-A*02:01 presenting MAGE-A10 9-mer peptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Simister, P.C, Border, E.C, Vieira, J.F, Pumphrey, N.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into engineering a T-cell receptor targeting MAGE-A10 with higher affinity and specificity for cancer immunotherapy.
J Immunother Cancer, 10, 2022
|
|
7YSZ
 
 | | Spiroplasma melliferum FtsZ bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-13 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3000412 Å) | | Cite: | Dynamics of interdomain rotation facilitates FtsZ filament assembly.
J.Biol.Chem., 300, 2024
|
|
8VFP
 
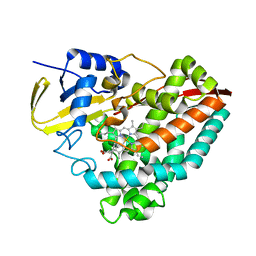 | |
7N55
 
 | | The crystal structure of the mutant I38T PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8PX7
 
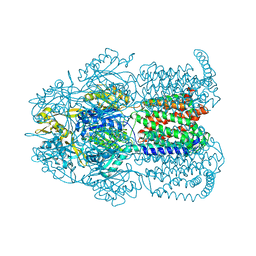 | | Structure of Bacterial Multidrug Efflux transporter AcrB, solved at wavelength 3.02 A | | Descriptor: | Multidrug efflux pump subunit AcrB | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Qu, F, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
5VJO
 
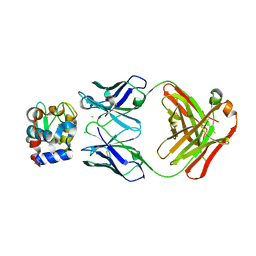 | |
