5DFI
 
 | | Human APE1 phosphorothioate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(OMC)P*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8JML
 
 | | Structure of Helicobacter pylori Soj protein mutant, D41A | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SpoOJ regulator (Soj) | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
6SDK
 
 | |
8JMK
 
 | | Structure of Helicobacter pylori Soj mutant, D41A bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
8JMJ
 
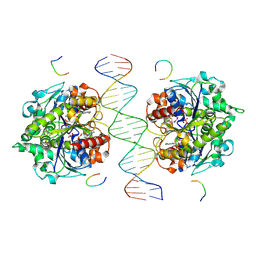 | | Structure of Helicobacter pylori Soj-DNA-Spo0J complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 52, 2024
|
|
7UNK
 
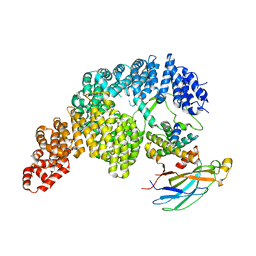 | | Structure of Importin-4 bound to the H3-H4-ASF1 histone-histone chaperone complex | | Descriptor: | Histone H3, Histone H4, Histone chaperone, ... | | Authors: | Bernardes, N.E, Chook, Y.M, Fung, H.Y.J, Chen, Z, Li, Y. | | Deposit date: | 2022-04-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure of IMPORTIN-4 bound to the H3-H4-ASF1 histone-histone chaperone complex.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5DOE
 
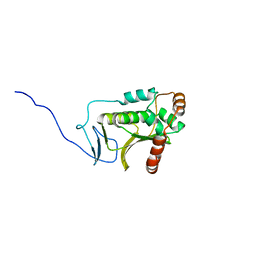 | | Crystal structure of the Human Cytomegalovirus UL53 (residues 72-292) | | Descriptor: | Virion egress protein UL31 homolog, ZINC ION | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
5DOB
 
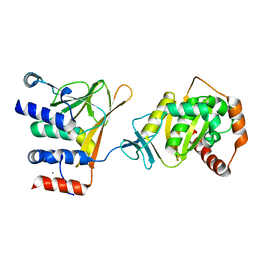 | | Crystal structure of the Human Cytomegalovirus Nuclear Egress Complex (NEC) | | Descriptor: | CALCIUM ION, Virion egress protein UL31 homolog, Virion egress protein UL34 homolog, ... | | Authors: | Lye, M.F, El Omari, K, Filman, D.J, Hogle, J.M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex.
Embo J., 34, 2015
|
|
4ACB
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH THE GTP ANALOGUE GPPNHP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
3H5N
 
 | | Crystal structure of E. coli MccB + ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MccB protein, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
4AC9
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS IN COMPLEX WITH GDP | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
4ACA
 
 | | CRYSTAL STRUCTURE OF TRANSLATION ELONGATION FACTOR SELB FROM METHANOCOCCUS MARIPALUDIS, APO FORM | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GUANOSINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Leibundgut, M, Frick, C, Thanbichler, M, Boeck, A, Ban, N. | | Deposit date: | 2011-12-14 | | Release date: | 2012-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Selenocysteine tRNA-Specific Elongation Factor Selb is a Structural Chimaera of Elongation and Initiation Factors.
Embo J., 24, 2005
|
|
5D98
 
 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
4C8V
 
 | | Xenopus RSPO2 Fu1-Fu2 crystal form I | | Descriptor: | R-SPONDIN-2 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8C
 
 | | mouse ZNRF3 ectodomain crystal form III | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8T
 
 | | Xenopus ZNRF3 ectodomain crystal form I | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C9V
 
 | |
4C9U
 
 | |
4C8P
 
 | |
8OEF
 
 | |
4AUI
 
 | | STRUCTURE AND FUNCTION OF THE PORB PORIN FROM DISSEMINATING N. GONORRHOEAE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Zeth, K, Kozjak-Pavlovic, V, Faulstich, M, Hurwitz, R, Kepp, O, Rudel, T. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Function of the Porb Porin from Disseminating Neisseria Gonorrhoeae.
Biochem.J., 449, 2013
|
|
7TJ5
 
 | | SthK closed state, cAMP-bound in the presence of POPA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TKT
 
 | | SthK closed state, cAMP-bound in the presence of detergent | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Rheinberger, J, Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TJ6
 
 | | SthK open state, cAMP-bound in the presence of POPA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Putative transcriptional regulator, ... | | Authors: | Schmidpeter, P.A, Nimigean, C.M. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Anionic lipids unlock the gates of select ion channels in the pacemaker family.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3H5R
 
 | | Crystal structure of E. coli MccB + Succinimide | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
