6AFU
 
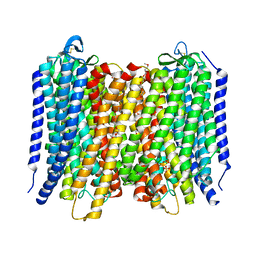 | | Proton pyrophosphatase-L555M mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
2YDQ
 
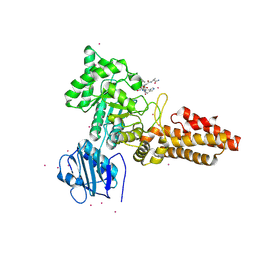 | | CpOGA D298N in complex with hOGA-derived O-GlcNAc peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIFUNCTIONAL PROTEIN NCOAT, CADMIUM ION, ... | | Authors: | Schimpl, M, Borodkin, V.S, Gray, L.J, van Aalten, D.M.F. | | Deposit date: | 2011-03-24 | | Release date: | 2012-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synergy of Peptide and Sugar in O-Glcnacase Substrate Recognition.
Chem.Biol., 19, 2012
|
|
2M8J
 
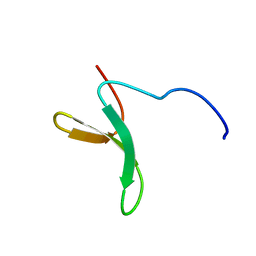 | | Structure of Pin1 WW domain phospho-mimic S16E | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Luh, L.M, Kirchner, D.K, Loehr, F, Haensel, R, Doetsch, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition.
J.Am.Chem.Soc., 135, 2013
|
|
6AD7
 
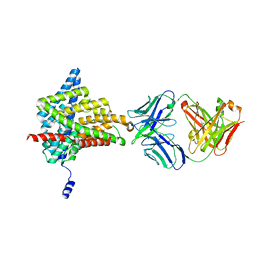 | | Crystal structure of the E148D mutant CLC-ec1 in 20 mM bromide | | Descriptor: | BROMIDE ION, H(+)/Cl(-) exchange transporter ClcA, antibody Fab fragment heavy chain, ... | | Authors: | Lim, H.-H, Park, K. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O88
 
 | | Crystal Structure of UDP-dependent glucosyltransferases (UGT) from Stevia rebaudiana in complex with UDP and rebaudioside A | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[alpha-L-allopyranosyl-(1->2)-[beta-D-mannopyranosyl-(1->3)]-beta-D-allopyranosyl]oxy}kauran-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for branched steviol glucoside biosynthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2YDR
 
 | | CpOGA D298N in complex with p53-derived O-GlcNAc peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, CELLULAR TUMOR ANTIGEN P53, ... | | Authors: | Schimpl, M, Borodkin, V.S, Gray, L.J, van Aalten, D.M.F. | | Deposit date: | 2011-03-24 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synergy of Peptide and Sugar in O-Glcnacase Substrate Recognition.
Chem.Biol., 19, 2012
|
|
6IE3
 
 | | Crystal structure of methyladenine demethylase | | Descriptor: | ETHANOL, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
6IED
 
 | | Crystal structure of heme A synthase from Bacillus subtilis | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Heme A synthase, ... | | Authors: | Niwa, S, Takeda, K, Kosugi, M, Tsutsumi, E, Miki, K. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of heme A synthase fromBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2YV6
 
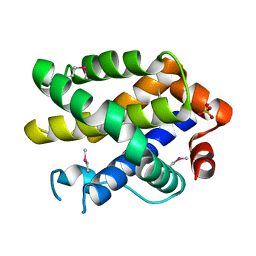 | | Crystal structure of human Bcl-2 family protein Bak | | Descriptor: | Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator.
J.Struct.Biol., 166, 2009
|
|
6ADC
 
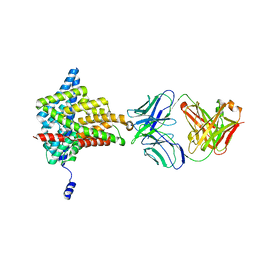 | |
2YM2
 
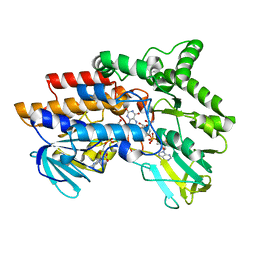 | | SNAPSHOTS OF ENZYMATIC BAEYER-VILLIGER CATALYSIS: OXYGEN ACTIVATION AND INTERMEDIATE STABILIZATION: Arg337Lys MUTANT REDUCED STATE WITH NADP | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PHENYLACETONE MONOOXYGENASE | | Authors: | Orru, R, Dudek, H.M, Martinoli, C, Torres Pazmino, D.E, Royant, A, Weik, M, Fraaije, M.W, Mattevi, A. | | Deposit date: | 2011-06-06 | | Release date: | 2011-06-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Snapshots of Enzymatic Baeyer-Villiger Catalysis: Oxygen Activation and Intermediate Stabilization.
J.Biol.Chem., 286, 2011
|
|
6AIY
 
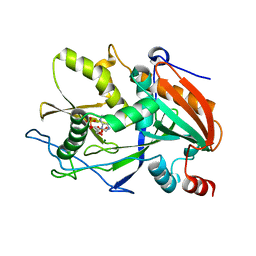 | |
2YVC
 
 | |
6O6G
 
 | | Co-crystal structure of Mcl1 with inhibitor | | Descriptor: | (3S)-5-(cyclobutylmethyl)-3-(2,4-dichlorophenyl)-2,3,4,5-tetrahydro-1,5-benzoxazepine-7-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Huang, X. | | Deposit date: | 2019-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AMG 176, a Selective MCL1 Inhibitor, Is Effective in Hematologic Cancer Models Alone and in Combination with Established Therapies.
Cancer Discov, 8, 2018
|
|
6IFF
 
 | | Crystal structure of M1 zinc metallopeptidase E323A mutant from Deinococcus radiodurans | | Descriptor: | SODIUM ION, TYROSINE, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Gaur, N.K, Makde, R.D. | | Deposit date: | 2018-09-20 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
6IIV
 
 | | Crystal structure of the human thromboxane A2 receptor bound to daltroban | | Descriptor: | 2-[4-[2-[(4-chlorophenyl)sulfonylamino]ethyl]phenyl]ethanoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | Fan, H, Zhao, Q, Wu, B. | | Deposit date: | 2018-10-07 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for ligand recognition of the human thromboxane A2receptor.
Nat. Chem. Biol., 15, 2019
|
|
6AFY
 
 | | Proton pyrophosphatase-E225S mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6O7W
 
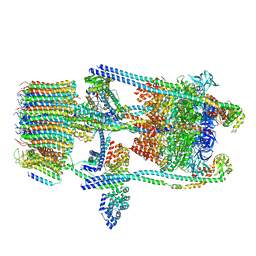 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 2 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6OEU
 
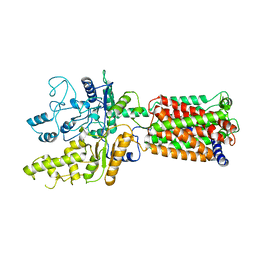 | | Structure of human Patched1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Protein patched homolog 1 | | Authors: | Qi, X, Li, X, Wang, J. | | Deposit date: | 2019-03-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of human Patched and its complex with native palmitoylated sonic hedgehog.
Nature, 560, 2018
|
|
6IKA
 
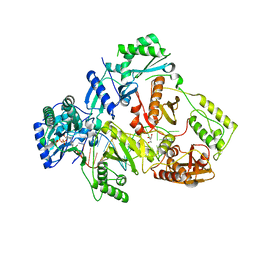 | | HIV-1 reverse transcriptase with Q151M/G112S/D113A/Y115F/F116Y/F160L/I159L:DNA:entecavir-triphosphate ternary complex | | Descriptor: | DNA/RNA (38-MER), GLYCEROL, HIV-1 RT p51 subunit, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Active-site deformation in the structure of HIV-1 RT with HBV-associated septuple amino acid substitutions rationalizes the differential susceptibility of HIV-1 and HBV against 4'-modified nucleoside RT inhibitors.
Biochem. Biophys. Res. Commun., 509, 2019
|
|
6OQS
 
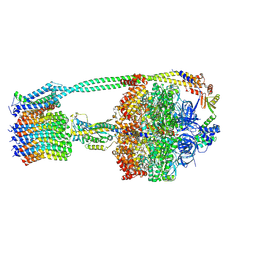 | | E. coli ATP synthase State 1b | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Stewart, A.G, Sobti, M, Walshe, J.L. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures provide insight into how E. coli F1FoATP synthase accommodates symmetry mismatch.
Nat Commun, 11, 2020
|
|
6OGD
 
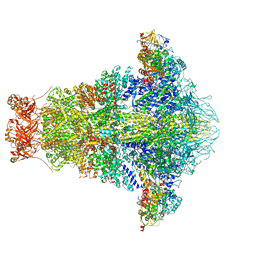 | | Cryo-EM structure of YenTcA in its prepore state | | Descriptor: | Chitinase 2, Toxin subunit YenA1, Toxin subunit YenA2 | | Authors: | Piper, S.J, Brillault, L, Box, J.K, Landsberg, M.J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin.
Nat Commun, 10, 2019
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6AKE
 
 | |
6AL2
 
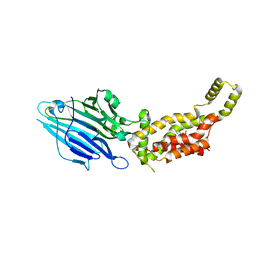 | | Crystal structure of E. coli YidC at 2.8 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Membrane protein insertase YidC | | Authors: | Tanaka, Y, Tsukazaki, T, Izumioka, A, Hamid, A.A, Fujii, A. | | Deposit date: | 2018-09-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8-angstrom crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
