3MXE
 
 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
8DWL
 
 | | Inhibitor-3:PP1 coexpressed complex | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase PPP1R11, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit, ... | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
5GNK
 
 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
7UOH
 
 | | PRMT5/MEP50 crystal structure with MTA and an achiral, class 1, non-atropisomeric inhibitor bound | | Descriptor: | (2M)-2-[(4M)-4-{4-(aminomethyl)-1-oxo-8-[(2R)-oxolan-2-yl]-1,2-dihydrophthalazin-6-yl}-1-methyl-1H-pyrazol-5-yl]-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and evaluation of achiral, non-atropisomeric 4-(aminomethyl)phthalazin-1(2H)-one derivatives as novel PRMT5/MTA inhibitors.
Bioorg.Med.Chem., 71, 2022
|
|
7Z6U
 
 | | Pim1 in complex with (E)-4-((6-amino-2-oxoindolin-3-ylidene)methyl)benzoic acid and Pimtide | | Descriptor: | 4-[(~{E})-(6-azanyl-2-oxidanylidene-1~{H}-indol-3-ylidene)methyl]benzoic acid, GLYCEROL, Isoform 1 of Serine/threonine-protein kinase pim-1, ... | | Authors: | Hochban, P.M.M, Heine, A, Diederich, W.E. | | Deposit date: | 2022-03-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Pose, duplicate, then elaborate: Steps towards increased affinity for inhibitors targeting the specificity surface of the Pim-1 kinase.
Eur.J.Med.Chem., 245, 2023
|
|
6H0U
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with a covalent [1,2,4]triazolo[1,5-a][1,3,5]triazine inhibitor | | Descriptor: | (2~{R})-3-[7-azanyl-5-(cyclohexylamino)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-2-yl]-2-cyano-propanamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marcovich, I, Demitri, N, De Zorzi, R, Storici, P. | | Deposit date: | 2018-07-10 | | Release date: | 2019-05-15 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Triazolotriazine-Based Dual GSK-3 beta /CK-1 delta Ligand as a Potential Neuroprotective Agent Presenting Two Different Mechanisms of Enzymatic Inhibition.
Chemmedchem, 14, 2019
|
|
8J51
 
 | |
8J50
 
 | |
5JAZ
 
 | | Structure of Plasmodium falciparum DXR in complex with a beta-substituted fosmidomycin analogue, LC51 and manganese | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplastic, FORMIC ACID, ... | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2016-04-12 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin.
Chemmedchem, 11, 2016
|
|
8T6V
 
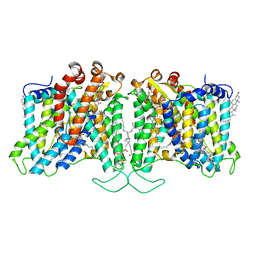 | | Cryo-EM structure of human Anion Exchanger 1 bound to 4,4'-Diisothiocyanatostilbene-2,2'-Disulfonic Acid (DIDS) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4'-Diisothiocyano-2,2'-stilbenedisulfonic acid, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T6U
 
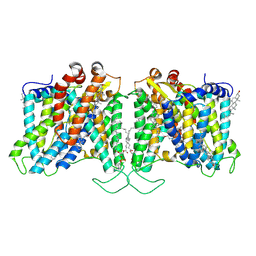 | | Cryo-EM structure of human Anion Exchanger 1 bound to Dipyridamole | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Capper, M.J, Zilberg, G, Mathiharan, Y.K, Yang, S, Stone, A.C, Wacker, D. | | Deposit date: | 2023-06-18 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Substrate binding and inhibition of the anion exchanger 1 transporter.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3NF7
 
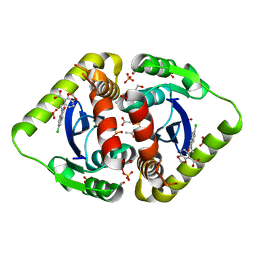 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
5FBN
 
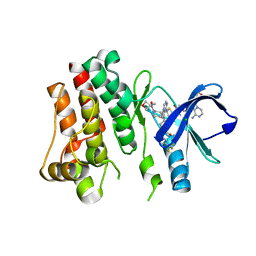 | | BTK kinase domain with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[8-azanyl-3-[(2~{S})-1-[4-(dimethylamino)butanoyl]pyrrolidin-2-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-(1,3-thiazol-2-yl)benzamide, 4-[8-azanyl-3-[(3~{R})-1-(3-methyloxetan-3-yl)carbonylpiperidin-3-yl]imidazo[1,5-a]pyrazin-1-yl]-~{N}-[4-(trifluoromethyl)pyridin-2-yl]benzamide, ... | | Authors: | Raaijmakers, H.C.A, Vu-Pham, D. | | Deposit date: | 2015-12-14 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of 8-Amino-imidazo[1,5-a]pyrazines as Reversible BTK Inhibitors for the Treatment of Rheumatoid Arthritis.
Acs Med.Chem.Lett., 7, 2016
|
|
8K66
 
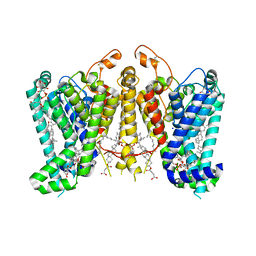 | | Cryo-EM structure of Oryza sativa HKT2;1 at 2.5 angstrom | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Wang, X, Shen, X, Qu, Y, Wang, C, Shen, H. | | Deposit date: | 2023-07-25 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural insights into ion selectivity and transport mechanisms of Oryza sativa HKT2;1 and HKT2;2/1 transporters.
Nat.Plants, 10, 2024
|
|
5JFU
 
 | | HIV-1 wild Type protease with GRL-007-14A (a Adamantane P1-Ligand with bis-THF in P2 and benzylamine in P1') | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-{benzyl[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
8SZ3
 
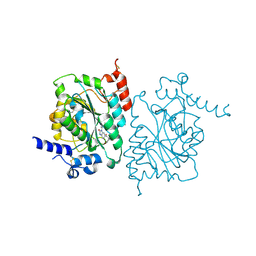 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 7j | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[(1S)-1-(5-bromopyridin-2-yl)ethyl]-3-[(2R)-3,3-dimethylbutan-2-yl]-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxamide, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-05-26 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
6RMR
 
 | |
8YD3
 
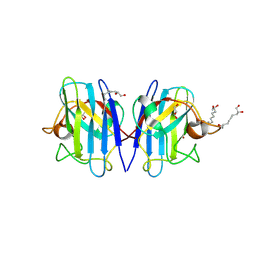 | | Crystal structure of human Cu-Zn Superoxide Dismutase 1 in complex with 1,2,10-Decanetriol | | Descriptor: | (2S)-decane-1,2,10-triol, GLYCEROL, Superoxide dismutase [Cu-Zn], ... | | Authors: | Aouti, S, Padmanabhan, B. | | Deposit date: | 2024-02-19 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of human Cu-Zn Superoxide Dismutase 1 in complex with 1,2,10-Decanetriol
To Be Published
|
|
3NF6
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NIY
 
 | | Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
3NF9
 
 | | Structural basis for a new mechanism of inhibition of HIV integrase identified by fragment screening and structure based design | | Descriptor: | 1,2-ETHANEDIOL, 5-[(6-chloro-2-oxo-2,3-dihydro-1H-indol-1-yl)methyl]-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S, Newman, J, Deadman, J.J, Rhodes, D. | | Deposit date: | 2010-06-10 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for a new mechanism of inhibition of HIV-1 integrase identified by fragment screening and structure-based design
ANTIVIR.CHEM.CHEMOTHER., 21, 2011
|
|
3NJ3
 
 | | Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
8RVK
 
 | | Maltodextrin phosphorylase (MalP) in complex with a alpha-1,2-cyclophellitol analogue | | Descriptor: | (3~{a}~{R},4~{R},5~{R},6~{R},7~{a}~{S})-6-(hydroxymethyl)-4,5-bis(oxidanyl)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-1,3-benzoxazol-2-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bennett, M, Ofman, T.P, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2024-02-01 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Conformational and Electronic Variations in 1,2- and 1,5a-Cyclophellitols and their Impact on Retaining alpha-Glucosidase Inhibition.
Chemistry, 30, 2024
|
|
3NU3
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
3NU6
 
 | |
