3WN1
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3S9D
 
 | | binary complex between IFNa2 and IFNAR2 | | Descriptor: | CHLORIDE ION, Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9999 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3KGW
 
 | |
3KHB
 
 | | Crystal structure of Escherichia coli AlkB with Co(II) and 2-OG | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB, COBALT (II) ION | | Authors: | Hollis, T, Holland, P.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and mutational analysis of Escherichia coli AlkB provides insight into substrate specificity and DNA damage searching.
Plos One, 5, 2010
|
|
3SCI
 
 | | Crystal structure of spike protein receptor-binding domain from a predicted SARS coronavirus human strain complexed with human receptor ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, CHLORIDE ION, Spike glycoprotein, ... | | Authors: | Wu, K, Peng, G, Wilken, M, Geraghty, R, Li, F. | | Deposit date: | 2011-06-07 | | Release date: | 2012-02-08 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus.
J.Biol.Chem., 287, 2012
|
|
3KLE
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3WPH
 
 | |
3WQ6
 
 | |
3SCU
 
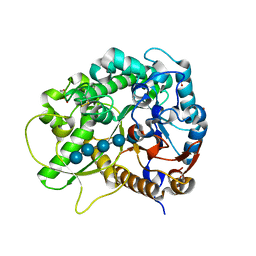 | | Crystal Structure of Rice BGlu1 E386G Mutant Complexed with Cellopentaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase 7, SULFATE ION, ... | | Authors: | Pengthaisong, S, Withers, S.G, Kuaprasert, B, Ketudat Cairns, J.R. | | Deposit date: | 2011-06-08 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural investigation of the basis for cellooligosaccharide synthesis by rice BGlu1 glycosynthases
to be published
|
|
3KM6
 
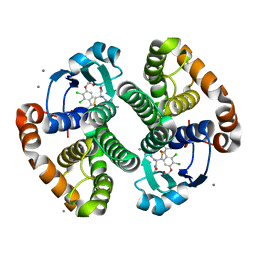 | |
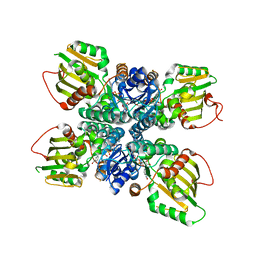
3OA2
 
 | |
5ZWV
 
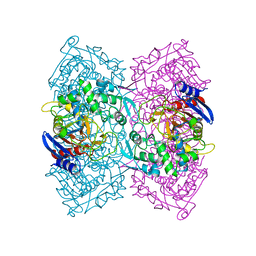 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29 | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W. | | Deposit date: | 2018-05-17 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
3KN1
 
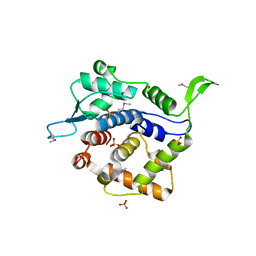 | | Crystal Structure of Golgi Phosphoprotein 3 N-term Truncation Variant | | Descriptor: | Golgi phosphoprotein 3, SULFATE ION | | Authors: | Schmitz, K.R, Bessman, N.J, Setty, T.G, Ferguson, K.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking.
J.Cell Biol., 187, 2009
|
|
3SE3
 
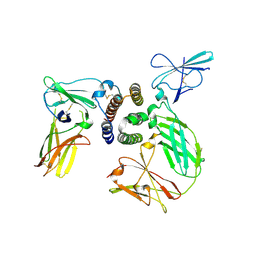 | | human IFNa2-IFNAR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon alpha 2b, Interferon alpha/beta receptor 1, ... | | Authors: | Thomas, C, Garcia, K.C. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.0001 Å) | | Cite: | Structural linkage between ligand discrimination and receptor activation by type I interferons.
Cell(Cambridge,Mass.), 146, 2011
|
|
3O0R
 
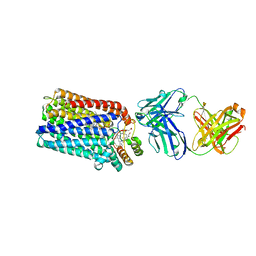 | | Crystal structure of nitric oxide reductase from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, FE (III) ION, HEME C, ... | | Authors: | Hino, T, Matsumoto, Y, Nagano, S, Sugimoto, H, Fukumori, Y, Murata, T, Iwata, S, Shiro, Y. | | Deposit date: | 2010-07-20 | | Release date: | 2010-12-29 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of biological N2O generation by bacterial nitric oxide reductase
Science, 330, 2010
|
|
3KOB
 
 | |
3O1R
 
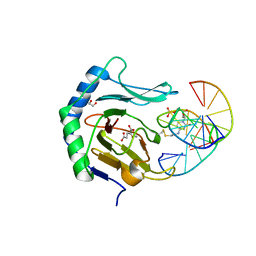 | | Iron-Catalyzed Oxidation Intermediates Captured in A DNA Repair Dioxygenase | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, DNA (5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Yi, C, Jia, G, Hou, G, Dai, Q, Zhang, W, Zheng, G, Jian, X, Yang, C.-G, Cui, Q, He, C. | | Deposit date: | 2010-07-21 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
Nature, 468, 2010
|
|
5ZZB
 
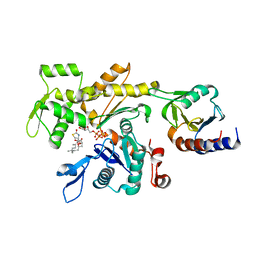 | | LokiProfilin2/Rabbit Actin Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Robinson, R.C, Akil, C. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genomes of Asgard archaea encode profilins that regulate actin.
Nature, 562, 2018
|
|
3ODK
 
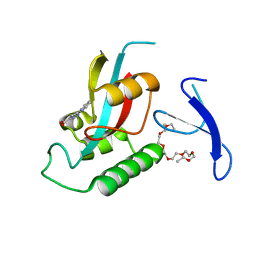 | | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-pyridin-2-yl-1H-pyrazole-5-carboxylic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3SGT
 
 | |
3SK2
 
 | |
4DOY
 
 | | Crystal structure of Dibenzothiophene desulfurization enzyme C | | Descriptor: | Dibenzothiophene desulfurization enzyme C, GLYCEROL | | Authors: | Liu, S, Zhang, C, Zhu, D, Gu, L. | | Deposit date: | 2012-02-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of DszC from Rhodococcus sp. XP at 1.79 angstrom
Proteins, 82, 2014
|
|
3WGU
 
 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state without oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
3RY2
 
 | | Wild-type core streptavidin-biotin complex at atomic resolution | | Descriptor: | BIOTIN, GLYCEROL, Streptavidin | | Authors: | Stenkamp, R.E, Le Trong, I, Stayton, P.S, Lybrand, T.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Streptavidin and its biotin complex at atomic resolution.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3WIZ
 
 | | Crystal structure of Bcl-xL in complex with compound 10 | | Descriptor: | 7-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]carbamoyl}-2-methylphenyl)-3-[3-(naphthalen-1-yloxy)propyl]pyrazolo[1,5-a]pyridine-2-carboxylic acid, Bcl-2-like protein 1, PHOSPHATE ION | | Authors: | Sogabe, S, Igaki, S, Hayano, Y. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of potent Mcl-1/Bcl-xL dual inhibitors by using a hybridization strategy based on structural analysis of target proteins.
J.Med.Chem., 56, 2013
|
|
