1UCN
 
 | | X-ray structure of human nucleoside diphosphate kinase A complexed with ADP at 2 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chen, Y, Gallois-Montbrun, S, Schneider, B, Veron, M, Morera, S, Deville-Bonne, D, Janin, J. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nucleotide Binding to Nucleoside Diphosphate Kinases: X-ray Structure of Human NDPK-A in Complex with ADP and Comparison to Protein Kinases
J.Mol.Biol., 332, 2003
|
|
8B0D
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, ALANINE, SULFATE ION, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
1UD2
 
 | | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) | | Descriptor: | GLYCEROL, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
8B1R
 
 | | RecBCD in complex with the phage protein gp5.9 | | Descriptor: | MAGNESIUM ION, Probable RecBCD inhibitor gp5.9, RecBCD enzyme subunit RecB, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
8BES
 
 | |
1UDU
 
 | | Crystal structure of Human Phosphodiesterase 5 complexed with tadalafil(Cialis) | | Descriptor: | 6-BENZO[1,3]DIOXOL-5-YL-2-METHYL-2,3,6,7,12,12A-HEXAHYDRO-PYRAZINO[1',2':1,6]PYRIDO[3,4-B]INDOLE-1,4-DIONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Sung, B.-J, Lee, J.I, Heo, Y.-S, Kim, J.H, Moon, J, Yoon, J.M, Hyun, Y.-L, Kim, E, Eum, S.J, Lee, T.G, Cho, J.M, Park, S.-Y, Lee, J.-O, Jeon, Y.H, Hwang, K.Y, Ro, S. | | Deposit date: | 2003-05-06 | | Release date: | 2004-05-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules
Nature, 425, 2003
|
|
8ASY
 
 | | SARS-CoV-2 Omicron BA.2.75 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-08-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75.
Cell Rep, 42, 2022
|
|
1U11
 
 | | PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) from the acidophile Acetobacter aceti | | Descriptor: | CITRIC ACID, PurE (N5-carboxyaminoimidazole Ribonucleotide Mutase) | | Authors: | Settembre, E.C, Chittuluru, J.R, Mill, C.P, Kappock, T.J, Ealick, S.E. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acidophilic adaptations in the structure of Acetobacter aceti N5-carboxyaminoimidazole ribonucleotide mutase (PurE).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U19
 
 | | Crystal Structure of Bovine Rhodopsin at 2.2 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEPTANE-1,2,3-TRIOL, MERCURY (II) ION, ... | | Authors: | Okada, T, Sugihara, M, Bondar, A.N, Elstner, M, Entel, P, Buss, V. | | Deposit date: | 2004-07-15 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure
J.Mol.Biol., 342, 2004
|
|
8BEQ
 
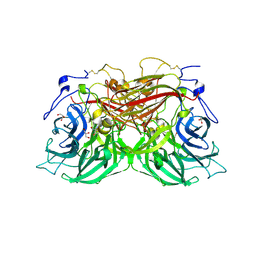 | | Structure of fructofuranosidase from Rhodotorula dairenensis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Insights into the Structure of the Highly Glycosylated Ffase from Rhodotorula dairenensis Enhance Its Biotechnological Potential.
Int J Mol Sci, 23, 2022
|
|
1UEG
 
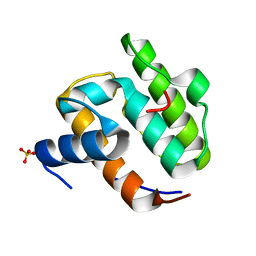 | |
8BEU
 
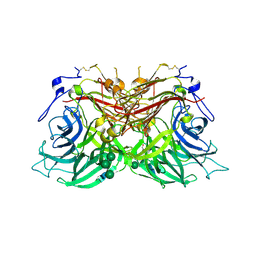 | |
8AW7
 
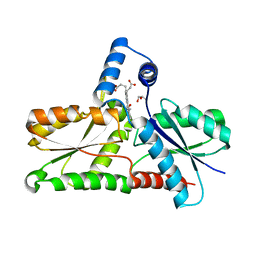 | | Structure of coproporphyrin III-LmCpfC R45L | | Descriptor: | Coproporphyrin III ferrochelatase, GLYCEROL, coproporphyrin III | | Authors: | Gabler, T, Hofbauer, S, Pfanzagl, V. | | Deposit date: | 2022-08-29 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Active site architecture of coproporphyrin ferrochelatase with its physiological substrate coproporphyrin III: Propionate interactions and porphyrin core deformation.
Protein Sci., 32, 2023
|
|
1UEN
 
 | | Solution Structure of The Third Fibronectin III Domain of Human KIAA0343 Protein | | Descriptor: | KIAA0343 protein | | Authors: | Miyamoto, K, Kigawa, T, Hayashi, F, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The Third Fibronectin III Domain of Human KIAA0343 Protein
To be Published
|
|
8B4O
 
 | | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum | | Descriptor: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome BD ubiquinol oxidase subunit I, Cytochrome bd-type quinol oxidase subunit II, ... | | Authors: | Grund, T.N, Kusumoto, T, Michel, H, Sakamoto, J, Safarian, S. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Cryo-EM structure of cytochrome bd oxidase from C. glutamicum
To be published
|
|
1UFK
 
 | | Crystal structure of TT0836 | | Descriptor: | TT0836 protein | | Authors: | Kaminishi, T, Sakai, H, Takemoto-Hori, C, Terada, T, Nakagawa, N, Maoka, N, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-31 | | Release date: | 2003-11-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of TT0836
To be Published
|
|
8BAL
 
 | |
1UFU
 
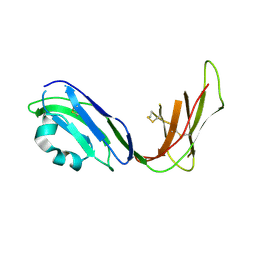 | | Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) | | Descriptor: | Immunoglobulin-like transcript 2 | | Authors: | Shiroishi, M, Amano, K, Rasubala, L, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and thermodynamic properties of the interaction between Immunoglobulin like transcript (ILT) and MHC class I
To be Published
|
|
1UG0
 
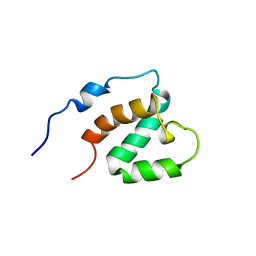 | | Solution structure of SURP domain in BAB30904 | | Descriptor: | splicing factor 4 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in BAB30904
To be Published
|
|
8BGN
 
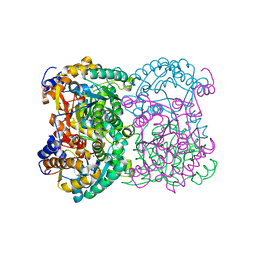 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Diacetylchitobiose deacetylase, ... | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
1U1K
 
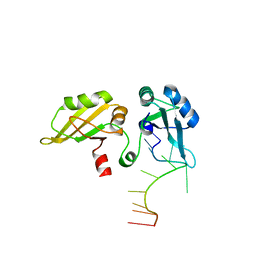 | |
8BHT
 
 | | ABCG2 turnover-1 state with tariquidar bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL, ... | | Authors: | Yu, Q, Kowal, J, Tajkhorshid, E, Locher, K.P. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Differential dynamics and direct interaction of bound ligands with lipids in multidrug transporter ABCG2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BD4
 
 | | TniQ-capped Tns-ATP-dsDNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*C)-3'), MAGNESIUM ION, ... | | Authors: | Querques, I, Schmitz, M, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
1U1N
 
 | |
8BC6
 
 | |
