3V0L
 
 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with a novel UDP-Gal derived inhibitor (2GW) | | Descriptor: | 5-phenyl-uridine-5'-alpha-d-galactosyl-diphosphate, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Palcic, M.M, Jorgensen, R. | | Deposit date: | 2011-12-08 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Base-modified Donor Analogues Reveal Novel Dynamic Features of a Glycosyltransferase.
J.Biol.Chem., 288, 2013
|
|
3UTH
 
 | | Crystal structure of Aspergillus fumigatus UDP galactopyranose mutase complexed with substrate UDP-Galp in reduced state | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Dhatwalia, R, Singh, H, Tanner, J.J. | | Deposit date: | 2011-11-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Small-angle X-ray Scattering Analysis of UDP-galactopyranose Mutase from the Pathogenic Fungus Aspergillus fumigatus.
J.Biol.Chem., 287, 2012
|
|
3K3V
 
 | |
3V1N
 
 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3K43
 
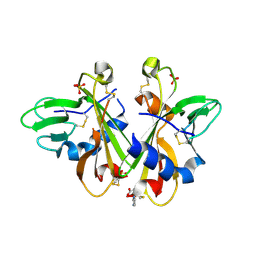 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
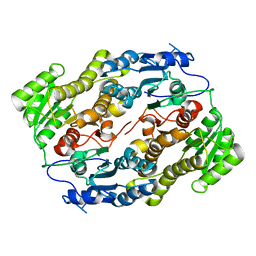
3V1T
 
 | |
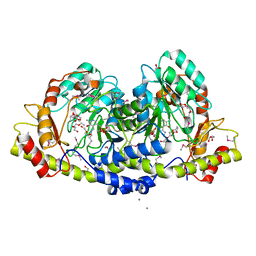
3JU7
 
 | |
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3UVQ
 
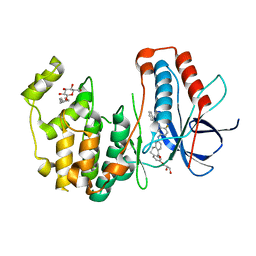 | | Human p38 MAP Kinase in Complex with a Dibenzosuberone Derivative | | Descriptor: | Mitogen-activated protein kinase 14, N-{5-[(7-{[(2R)-2,3-dihydroxypropyl]oxy}-5-oxo-10,11-dihydro-5H-dibenzo[a,d][7]annulen-2-yl)amino]-2-fluorophenyl}benzamide, octyl beta-D-glucopyranoside | | Authors: | Mayer-Wrangowski, S.C, Richters, A, Gruetter, C, Rauh, D. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dibenzosuberones as p38 mitogen-activated protein kinase inhibitors with low ATP competitiveness and outstanding whole blood activity.
J.Med.Chem., 56, 2013
|
|
3V4S
 
 | | Crystal Structure of ADP-ATP complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-12-15 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3V5Q
 
 | | Discovery of a selective TRK Inhibitor with efficacy in rodent cancer tumor models | | Descriptor: | 1-(3-{[(3Z)-2-oxo-3-(1H-pyrrol-2-ylmethylidene)-2,3-dihydro-1H-indol-6-yl]amino}phenyl)-3-[3-(trifluoromethyl)phenyl]urea, CHLORIDE ION, NT-3 growth factor receptor | | Authors: | Kreusch, A. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Discovery of GNF-5837, a Selective TRK Inhibitor with Efficacy in Rodent Cancer Tumor Models.
ACS Med Chem Lett, 3, 2012
|
|
3V1K
 
 | |
3V5Y
 
 | | Structure of FBXL5 hemerythrin domain, P2(1) cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3JVO
 
 | | Crystal structure of bacteriophage HK97 gp6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Gp6 | | Authors: | Lam, R, Tuite, A, Battaile, K.P, Edwards, A.M, Maxwell, K.L, Chirgadze, N.Y. | | Deposit date: | 2009-09-17 | | Release date: | 2009-11-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins.
J.Mol.Biol., 395, 2010
|
|
3JXE
 
 | | Crystal structure of Pyrococcus horikoshii tryptophanyl-tRNA synthetase in complex with TrpAMP | | Descriptor: | SULFATE ION, TRYPTOPHANYL-5'AMP, Tryptophanyl-tRNA synthetase | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Yang, B, Ding, J. | | Deposit date: | 2009-09-19 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of P. horikoshii tryptophanyl-tRNA synthetase and structure-based phylogenetic analysis suggest an archaeal origin of tryptophanyl-tRNA synthetase
To be Published
|
|
3V8A
 
 | | Thaumatin by Classical Hanging Drop Vapour Diffusion after 18.1 MGy X-Ray dose at ESRF ID29 beamline (Worst Case) | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Belmonte, L, Scudieri, D, Tripathi, S, Pechkova, E, Nicolini, C. | | Deposit date: | 2011-12-22 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Langmuir-Blodgett nanotemplate and radiation resistance in protein crystals: state of the art.
CRIT.REV.EUKARYOT.GENE EXPR., 22, 2012
|
|
3V9A
 
 | |
3JZJ
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
3K08
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-NTPP, Na+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Potassium channel protein NaK, SODIUM ION | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K0H
 
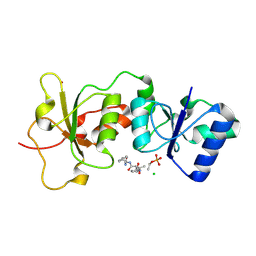 | | The crystal structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3V9F
 
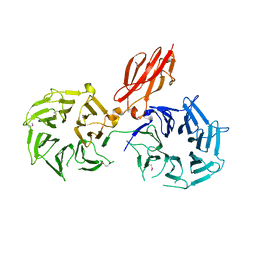 | |
3V9J
 
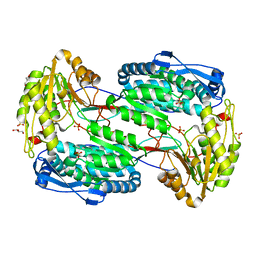 | |
3K0Y
 
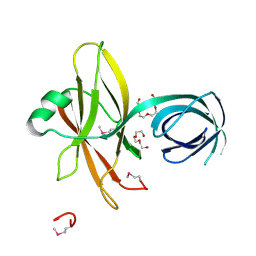 | |
3K15
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3VAI
 
 | | Structure of U2AF65 variant with BrU3C5 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DNA 5'-D(*UP*UP*(BRU)P*UP*CP*UP*U)-3', GLYCEROL, ... | | Authors: | Jenkins, J.L, Frato, K.H, Kielkopf, C.L. | | Deposit date: | 2011-12-29 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | U2AF65 adapts to diverse pre-mRNA splice sites through conformational selection of specific and promiscuous RNA recognition motifs.
Nucleic Acids Res., 41, 2013
|
|
