5D7A
 
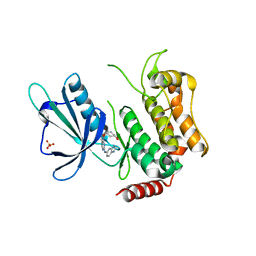 | | Crystal structure of the kinase domain of TRAF2 and NCK-interacting protein kinase with NCB-0846 | | Descriptor: | SULFATE ION, TRAF2 and NCK-interacting protein kinase, cis-4-{[2-(1H-benzimidazol-5-ylamino)quinazolin-8-yl]oxy}cyclohexanol | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-08-13 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
5CWZ
 
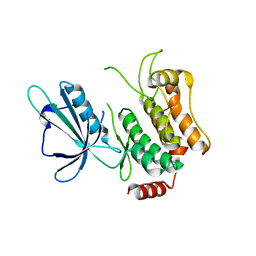 | |
5AX9
 
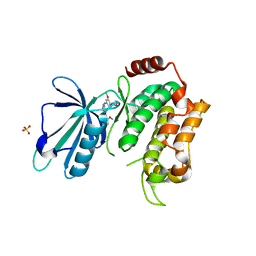 | | Crystal structure of the kinase domain of human TRAF2 and NCK-interacting protein kinase in complex with compund 9 | | Descriptor: | 4-methoxy-3-[2-[(3-methoxy-4-morpholin-4-yl-phenyl)amino]pyridin-4-yl]benzenecarbonitrile, SULFATE ION, TRAF2 and NCK-interacting protein kinase | | Authors: | Ohbayashi, N, Kukimoto-Niino, M, Yamada, T, Shirouzu, M. | | Deposit date: | 2015-07-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | TNIK inhibition abrogates colorectal cancer stemness
Nat Commun, 7, 2016
|
|
6LKC
 
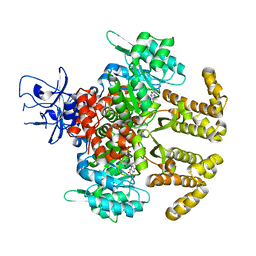 | | Crystal structure of PfaD from Shewanella piezotolerans in complex with FMN | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Zhang, M.L, Li, Q, Meng, S.S, Guo, L.J, He, L, Huang, J.Z, Li, L, Zhang, H.D. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Trans -Acting Enoyl Reductase in the Biosynthesis of Long-Chain Polyunsaturated Fatty Acids in Shewanella piezotolerans .
J.Agric.Food Chem., 69, 2021
|
|
5U9K
 
 | |
5U9A
 
 | |
5U9J
 
 | | Crystal structure of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with geranyl geranyl pyrophoshate | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), GERAN-8-YL GERAN, ISOPROPYL ALCOHOL, ... | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U9I
 
 | | Crystal structure of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5V35
 
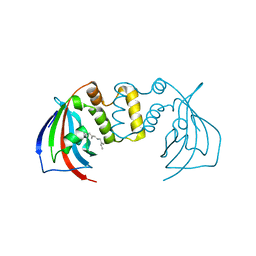 | | Crystal structure of V71F mutant of the FKBP domain of human aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1) complexed with S-farnesyl-L-cysteine methyl ester | | Descriptor: | Aryl hydrocarbon receptor-interacting protein-like 1 (AIPL1), FARNESYL | | Authors: | Yadav, R.P, Gakhar, L, Liping, Y, Artemyev, N.O. | | Deposit date: | 2017-03-06 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique structural features of the AIPL1-FKBP domain that support prenyl lipid binding and underlie protein malfunction in blindness.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | Descriptor: | Striatin-3 | | Authors: | Chen, C, Shi, Z, Zhou, Z. | | Deposit date: | 2013-10-13 | | Release date: | 2014-02-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
4Z38
 
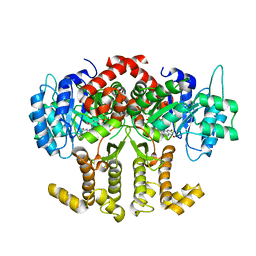 | |
4UT5
 
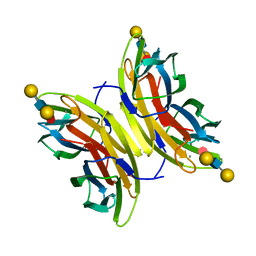 | | Crystal structure of the LecB lectin from Pseudomonas aeruginosa strain PA7 in complex with lewis a tetrasaccharide | | Descriptor: | CALCIUM ION, LECB LECTIN, beta-D-galactopyranose, ... | | Authors: | Boukerb, A.M, Decor, A, Tabaroni, R, Varrot, A, Debentzmann, S, Vidal, S, Imberty, A, Cournoyer, B. | | Deposit date: | 2014-07-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genomic Rearrangements and Functional Diversification of Leca and Lecb Lectin-Coding Regions Impacting the Efficacy of Glycomimetics Directed Against Pseudomonas Aeruginosa.
Front.Microbiol., 7, 2016
|
|
5AWR
 
 | |
6E31
 
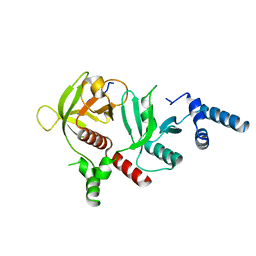 | | Crystal structure of RIAM in an autoinhibited configuration. | | Descriptor: | Rap1-interacting adapter molecule | | Authors: | Chang, Y.C, Su, W, Zhang, H, Huang, Q, Philips, M.R, Wu, J. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for autoinhibition of RIAM regulated by FAK in integrin activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6U46
 
 | |
6U79
 
 | |
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
5CE7
 
 | | Structure of a non-canonical CID of Ctk3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CTD kinase subunit gamma | | Authors: | Muehlbacher, W, Mayer, A, Sun, M, Remmert, M, Cheung, A.C, Niesser, J, Soeding, J, Cramer, P. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Ctk3, a subunit of the RNA polymerase II CTD kinase complex, reveals a noncanonical CTD-interacting domain fold.
Proteins, 83, 2015
|
|
5D94
 
 | | Crystal structure of LC3-LIR peptide complex | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Peptide from FYVE and coiled-coil domain-containing protein 1 | | Authors: | Takagi, K, Mizushima, T, Johansen, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | FYCO1 Contains a C-terminally Extended, LC3A/B-preferring LC3-interacting Region (LIR) Motif Required for Efficient Maturation of Autophagosomes during Basal Autophagy
J.Biol.Chem., 290, 2015
|
|
6W3M
 
 | |
5AWS
 
 | |
5AWT
 
 | |
5DF6
 
 | | Crystal structure of PTPN11 tandem SH2 domains in complex with a TXNIP peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11, UNKNOWN ATOM OR ION, txnip | | Authors: | Dong, A, Li, W, Tempel, W, Liu, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for the regulatory role of the PPxY motifs in the thioredoxin-interacting protein TXNIP.
Biochem.J., 473, 2016
|
|
5AWU
 
 | |
6DSI
 
 | |
