5PX7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 31) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PXN
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 47) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PY4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 64) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PYM
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 82) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PZ0
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 96) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PZG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SP100 after initial refinement with no ligand modelled (structure 112) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6I7R
 
 | |
7OFL
 
 | | Crystal structure of the sesquiterpene synthase Copu9 from coniophora puteana in complex with alendronate | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, MAGNESIUM ION, ... | | Authors: | Dimos, N, Loll, B. | | Deposit date: | 2021-05-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Biotechnological potential and initial characterization of two novel sesquiterpene synthases from Basidiomycota Coniophora puteana for heterologous production of delta-cadinol.
Microb Cell Fact, 21, 2022
|
|
6I7Q
 
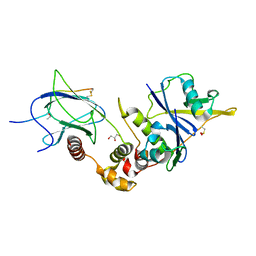 | |
2MR8
 
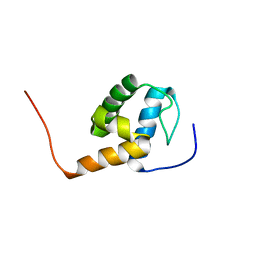 | |
4LHT
 
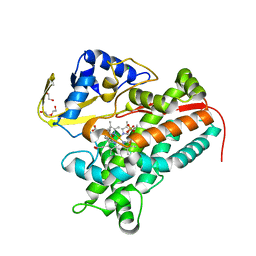 | | Crystal Structure of P450cin Y81F mutant, crystallized in 3 mM 1,8-cineole | | Descriptor: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Madrona, Y, Poulos, T.L. | | Deposit date: | 2013-07-01 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | P450cin active site water: implications for substrate binding and solvent accessibility.
Biochemistry, 52, 2013
|
|
2KXG
 
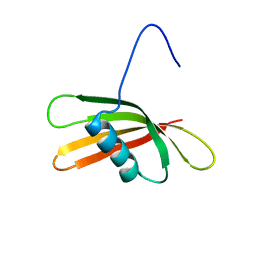 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
2MR7
 
 | |
7AO2
 
 | | Crystal structure of CotB2 variant W288G in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
7AO3
 
 | | Crystal structure of CotB2 variant F149L in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
7AO0
 
 | | Crystal structure of CotB2 variant F107A in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
7AO1
 
 | | Crystal structure of CotB2 variant W288F in complex with alendronate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, CHLORIDE ION, ... | | Authors: | Dimos, N, Driller, R, Loll, B. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Impression of a Nonexisting Catalytic Effect: The Role of CotB2 in Guiding the Complex Biosynthesis of Cyclooctat-9-en-7-ol.
J.Am.Chem.Soc., 142, 2020
|
|
2F92
 
 | | Crystal structure of human FPPS in complex with alendronate | | Descriptor: | 4-AMINO-1-HYDROXYBUTANE-1,1-DIYLDIPHOSPHONATE, Farnesyl Diphosphate Synthase, PHOSPHATE ION, ... | | Authors: | Rondeau, J.-M, Bitsch, F, Bourgier, E, Geiser, M, Hemmig, R, Kroemer, M, Lehmann, S, Ramage, P, Rieffel, S, Strauss, A, Green, J.R, Jahnke, W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the exceptional in vivo efficacy of bisphosphonate drugs.
Chemmedchem, 1, 2006
|
|
2FH0
 
 | |
7MBO
 
 | | FACTOR XIA (PICHIA PASTORIS; C500S [C122S]) IN COMPLEX WITH THE INHIBITOR Milvexian (BMS-986177), IUPAC NAME:(6R,10S)-10-{4-[5-chloro-2-(4-chloro-1H-1,2,3-triazol-1-yl)phenyl]-6- oxopyrimidin-1(6H)-yl}-1-(difluoromethyl)-6-methyl-1,4,7,8,9,10-hexahydro-15,11- (metheno)pyrazolo[4,3-b][1,7]diazacyclotetradecin-5(6H)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor XIa light chain, Milvexian | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.924 Å) | | Cite: | Discovery of Milvexian, a High-Affinity, Orally Bioavailable Inhibitor of Factor XIa in Clinical Studies for Antithrombotic Therapy.
J.Med.Chem., 65, 2022
|
|
3D2T
 
 | |
3NT9
 
 | |
6ITK
 
 | |
5KXT
 
 | | Hen Egg White Lysozyme at 278K, Data set 5 | | Descriptor: | Lysozyme C, SODIUM ION | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
5KVW
 
 | | T. danielli thaumatin at 100K, Data set 1 | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Russi, S, Gonzalez, A, Kenner, L.R, Keedy, D.A, Fraser, J.S, van den Bedem, H. | | Deposit date: | 2016-07-15 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Conformational variation of proteins at room temperature is not dominated by radiation damage.
J Synchrotron Radiat, 24, 2017
|
|
