6DD3
 
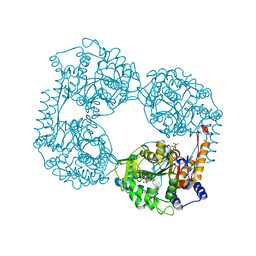 | | Crystal structure of the double mutant (D52N/D407A) of NT5C2-537X in the active state | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DD4
 
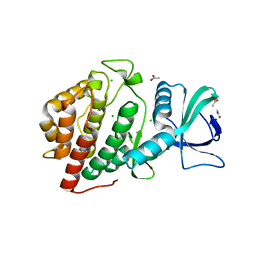 | | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-dipropyl-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, Santiago, A.S, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-09 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N,N-dipropyl-dihydropteridine inhibitor
To Be Published
|
|
6DD5
 
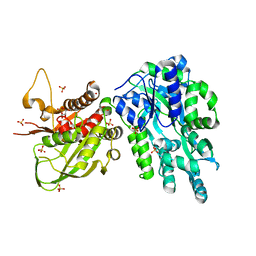 | | Crystal Structure of the Cas6 Domain of Marinomonas mediterranea MMB-1 Cas6-RT-Cas1 Fusion Protein | | Descriptor: | GLYCEROL, MMB-1 Cas6 Fused to Maltose Binding Protein,CRISPR-associated endonuclease Cas1, SULFATE ION, ... | | Authors: | Stamos, J.L, Mohr, G, Silas, S, Makarova, K.S, Markham, L.M, Yao, J, Lucas-Elio, P, Sanchez-Amat, A, Fire, A.Z, Koonin, E.V, Lambowitz, A.M. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition.
Mol. Cell, 72, 2018
|
|
6DD6
 
 | |
6DD7
 
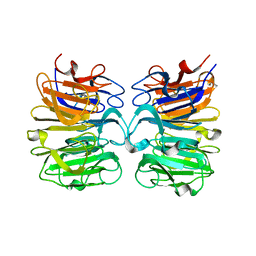 | |
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6DDA
 
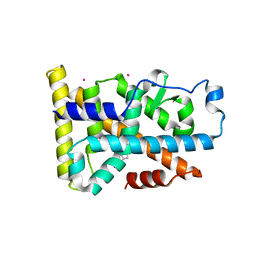 | | Nurr1 Covalently Modified by a Dopamine Metabolite | | Descriptor: | 5-hydroxy-1,2-dihydro-6H-indol-6-one, BROMIDE ION, Nuclear receptor subfamily 4 group A member 2, ... | | Authors: | Bruning, J.M, Wang, Y, Otrabella, F, Boxue, T, Liu, H, Bhattacharya, P, Guo, S, Holton, J.M, Fletterick, R.J, Jacobson, M.P, England, P.M. | | Deposit date: | 2018-05-09 | | Release date: | 2019-03-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Covalent Modification and Regulation of the Nuclear Receptor Nurr1 by a Dopamine Metabolite.
Cell Chem Biol, 26, 2019
|
|
6DDB
 
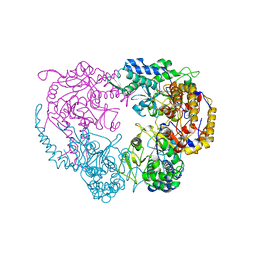 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the basal state, Northeast Structural Genomics Consortium Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDC
 
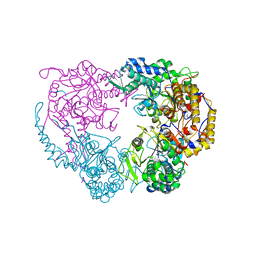 | | Crystal structure of the single mutant (D52N) of NT5C2-537X in the basal state, Northeast Structural Genomics Consortium Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDD
 
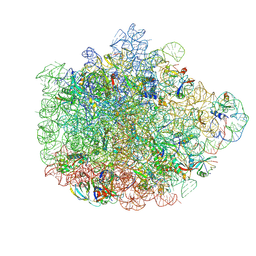 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-5 | | Descriptor: | 2,2-dichloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | Authors: | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6DDE
 
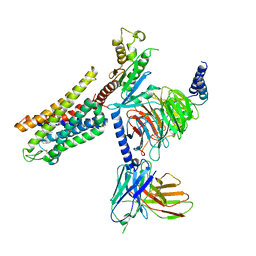 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Zhang, Y, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
6DDF
 
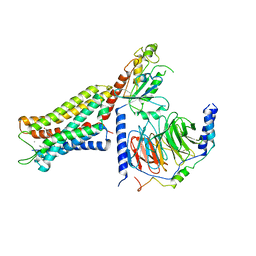 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
6DDG
 
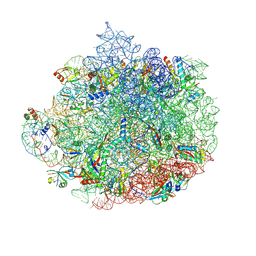 | | Structure of the 50S ribosomal subunit from Methicillin Resistant Staphylococcus aureus in complex with the oxazolidinone antibiotic LZD-6 | | Descriptor: | 2-chloro-N-({(5S)-3-[3-fluoro-4-(morpholin-4-yl)phenyl]-2-oxo-1,3-oxazolidin-5-yl}methyl)acetamide, 23S rRNA, 50S ribosomal protein L13, ... | | Authors: | Belousoff, M.J, Venugopal, H, Bamert, R.S, Lithgow, T. | | Deposit date: | 2018-05-10 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | cryoEM-Guided Development of Antibiotics for Drug-Resistant Bacteria.
ChemMedChem, 14, 2019
|
|
6DDH
 
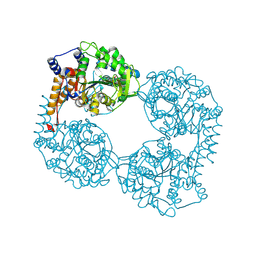 | | Crystal structure of the double mutant (D52N/R367Q) of NT5C2-537X in the active state, Northeast Structural Genomics Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, INOSINIC ACID | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDI
 
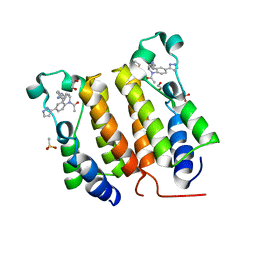 | | Crystal Structure of the human BRD2 BD1 bromodomain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
6DDJ
 
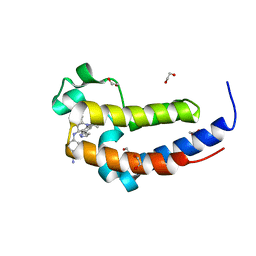 | | Crystal Structure of the human BRD2 BD2 bromodimain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
6DDK
 
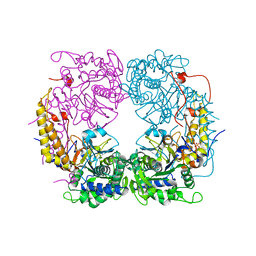 | | Crystal structure of the double mutant (D52N/R367Q) of the full-length NT5C2 in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDL
 
 | | Crystal structure of the single mutant (D52N) of NT5C2-Q523X in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDM
 
 | | Crystal Structure Analysis of the Epitope of an Anti-MICA Antibody | | Descriptor: | Anti-MICA Fab fragment heavy chain clone 1D5, Anti-MICA Fab fragment light chain clone 1D5, MHC class I polypeptide-related sequence A | | Authors: | Matsumoto, M.L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies.
MAbs, 11, 2019
|
|
6DDN
 
 | |
6DDO
 
 | | Crystal structure of the single mutant (D52N) of the full-length NT5C2 in the basal state | | Descriptor: | Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDP
 
 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
6DDQ
 
 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the basal state | | Descriptor: | 1,2-ETHANEDIOL, Cytosolic purine 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDR
 
 | | Crystal Structure Analysis of the Epitope of an Anti-MICA Antibody | | Descriptor: | Anti-MICA Fab fragment heavy chain clone 13A9, Anti-MICA Fab fragment light chain clone 13A9, GLYCEROL, ... | | Authors: | Matsumoto, M.L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies.
MAbs, 11, 2019
|
|
