8OIY
 
 | |
5TWA
 
 | | Crystal structure of Geodia cydonium BHP2 in complex with Lubomirskia baicalensis Bak-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAK-2 protein, ... | | Authors: | Caria, S, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2016-11-12 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into an evolutionarily ancient programmed cell death regulator - the crystal structure of marine sponge BHP2 bound to LB-Bak-2.
Cell Death Dis, 8, 2017
|
|
7VK5
 
 | |
7YZS
 
 | |
5CD5
 
 | |
6N7Q
 
 | |
7NG6
 
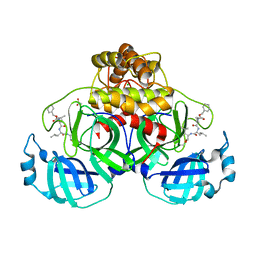 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1 in absence of DTT. | | Descriptor: | 3C-like proteinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
8OMW
 
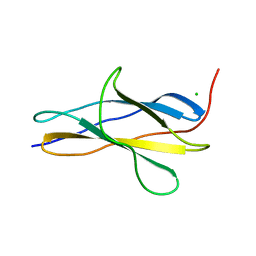 | | Crystal structure of the titin domain Fn3-20 | | Descriptor: | CHLORIDE ION, Titin | | Authors: | Rees, M, Gautel, M. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
7NE2
 
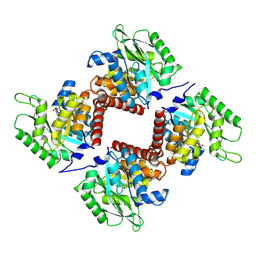 | | Crystal structure of class I SFP aldolase YihT from Salmonella enterica with SFP/ DHAP (Schiff base complex with active site Lys193) | | Descriptor: | (2~{S},3~{S},4~{R})-2,3,4,5-tetrakis(oxidanyl)-6-phosphonooxy-hexane-1-sulfonic acid, Sulfofructosephosphate aldolase, [(~{E})-2,3-bis(oxidanyl)prop-1-enyl] dihydrogen phosphate | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
5CGC
 
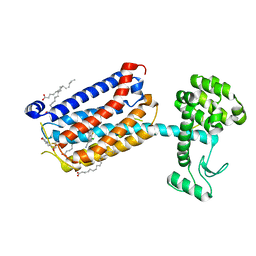 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5, ... | | Authors: | Christopher, J.A, Aves, S.J, Bennett, K.A, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Okrasa, K, Serrano-Vega, M.J, Tehan, B.G, Wiggin, G.R, Congreve, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
J.Med.Chem., 58, 2015
|
|
7VBU
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 5 | | Descriptor: | 8-cyclopropyl-2-methyl-9H-pyrido[2,3-b]indole, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
7ZC3
 
 | | Crystal structure of human copper chaperone Atox1 bound to zinc ion by CxxC motif | | Descriptor: | Copper transport protein ATOX1, SULFATE ION, ZINC ION | | Authors: | Mangini, V, Belviso, B.D, Arnesano, F, Caliandro, R. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Copper Chaperone ATOX1 Bound to Zinc Ion.
Biomolecules, 12, 2022
|
|
8UQX
 
 | | Round 18 Arylesterase Variant of Apo-Phosphotriesterase Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Phosphotriesterase variant PTE-R18 | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
5CGM
 
 | | Structure of Mycobacterium thermoresistibile GlgE in complex with maltose at 1.95A resolution | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase, CHLORIDE ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Maranha, A, Empadinhas, N, Blundell, T.L. | | Deposit date: | 2015-07-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mycobacterium thermoresistibile GlgE defines novel conformational states that contribute to the catalytic mechanism.
Sci Rep, 5, 2015
|
|
8OS3
 
 | | Crystal structure of the titin domain Fn3-11 | | Descriptor: | Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
5CDO
 
 | | 3.15A structure of QPT-1 with S.aureus DNA gyrase and DNA | | Descriptor: | (2R,4S,4aS)-4',6'-dihydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidin]-2'-one, (2R,4S,4aS,5R)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, (2R,4S,4aS,5S)-6'-hydroxy-2,4-dimethyl-8-nitro-1,2,4,4a-tetrahydro-2'H,6H-spiro[1,4-oxazino[4,3-a]quinoline-5,5'-pyrimidine]-2',4'(3'H)-dione, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
8OT5
 
 | | Crystal structure of the titin domain Fn3-85 | | Descriptor: | CHLORIDE ION, SODIUM ION, Titin | | Authors: | Nikoopour, R, Rees, M, Gautel, M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure determination and analysis of titin A-band fibronectin type III domains provides insights for disease-linked variants and protein oligomerisation.
J.Struct.Biol., 215, 2023
|
|
8UVM
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with covalent inhibitor Jun11313 | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Ansari, A, Tan, B, Chopra, A, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-03 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
5CHC
 
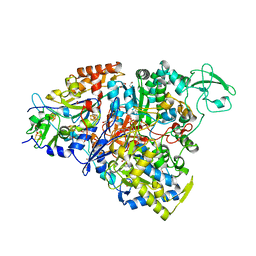 | | Crystal structure of the perchlorate reductase PcrAB - substrate analog SeO3 bound - from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BISELENITE ION, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-07-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
6N8S
 
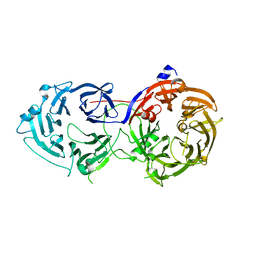 | |
7YZU
 
 | | Crystal structure of the sulfoquinovosyl binding protein SmoF complexed with SQMe | | Descriptor: | Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-methoxy-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Snow, A.J.D, Davies, G.J. | | Deposit date: | 2022-02-21 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The sulfoquinovosyl glycerol binding protein SmoF binds and accommodates plant sulfolipids.
Curr Res Struct Biol, 4, 2022
|
|
7VBX
 
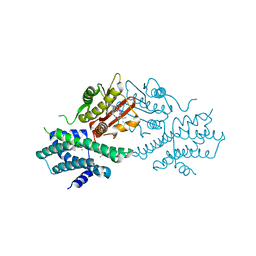 | | Crystal structure of human pyruvate dehydrogenase kinase 2 in complex with compound 20 | | Descriptor: | (3~{S})-3-[5-(8-cyclopropyl-2-methyl-9~{H}-pyrido[2,3-b]indol-3-yl)-1,3,4-oxadiazol-2-yl]-4-methyl-~{N}-[(1~{R})-1-phenylethyl]pentanamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Orita, T, Doi, S, Iwanaga, T, Fujishima, A, Adachi, T. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug design of novel and highly potent pyruvate dehydrogenase kinase inhibitors.
Bioorg.Med.Chem., 52, 2021
|
|
7Z79
 
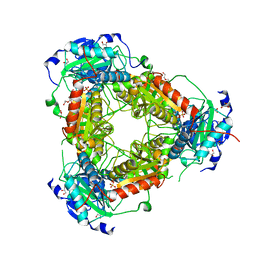 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
5TTO
 
 | | X-ray crystal structure of PPARgamma in complex with SR1643 | | Descriptor: | 4-bromo-N-{3,5-dichloro-4-[(quinolin-3-yl)oxy]phenyl}-2,5-difluorobenzene-1-sulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Bruning, J.B, Frkic, R.L, Griffin, P, Kamenecka, T, Abell, A. | | Deposit date: | 2016-11-04 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Structure-Activity Relationship of 2,4-Dichloro-N-(3,5-dichloro-4-(quinolin-3-yloxy)phenyl)benzenesulfonamide (INT131) Analogs for PPAR gamma-Targeted Antidiabetics.
J. Med. Chem., 60, 2017
|
|
6N9B
 
 | | FtsY-NG ultra high-resolution | | Descriptor: | ACETATE ION, Signal recognition particle receptor FtsY | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
