2RUK
 
 | |
3ILO
 
 | | Crystal structure of E. coli HPPK(D97A) in complex with MgAMPCPP and 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
3IP0
 
 | |
6MSC
 
 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3-methyl-1-(3-methylpyridin-4-yl)-3H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
8HCG
 
 | | Crystal structure of mTREX1-dAMP complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
6IJH
 
 | | Crystal structure of PDE10 in complex with inhibitor AF-399/14387019 | | Descriptor: | 2-[2-(4-phenyl-5-sulfanylidene-4,5-dihydro-1H-1,2,4-triazol-3-yl)ethyl]-1H-benzo[de]isoquinoline-1,3(2H)-dione, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.Y, Yu, Y.F, Zhang, C, Wu, D, Wu, Y, Luo, H.B. | | Deposit date: | 2018-10-10 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Validation of Phosphodiesterase-10 as a Novel Target for Pulmonary Arterial Hypertension via Highly Selective and Subnanomolar Inhibitors.
J. Med. Chem., 62, 2019
|
|
6CFB
 
 | | Isolation, Characterization, and Synthesis of the Barrettides: Disulfide-Containing Peptides from the Marine Sponge Geodia barretti | | Descriptor: | barrettide A | | Authors: | Rosengren, K.J, Carstens, B.B, Clark, R.J, Goransson, U. | | Deposit date: | 2018-02-14 | | Release date: | 2018-03-21 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Isolation, Characterization, and Synthesis of the Barrettides: Disulfide-Containing Peptides from the Marine Sponge Geodia barretti.
J. Nat. Prod., 78, 2015
|
|
6CEI
 
 | |
6CCW
 
 | |
6IFY
 
 | | Type III-A Csm complex, Cryo-EM structure of Csm-CTR1 | | Descriptor: | CTR1, Type III-A CRISPR-associated RAMP protein Csm3, Type III-A CRISPR-associated RAMP protein Csm4, ... | | Authors: | You, L, Ma, J, Wang, J, Zhang, X, Wang, Y. | | Deposit date: | 2018-09-21 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6MSA
 
 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 7,8-dimethoxy-1-methyl-2H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
3ILJ
 
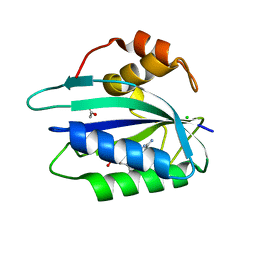 | | Crystal structure of E. coli HPPK(D95A) in complex with MgAMPCPP | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
3J8A
 
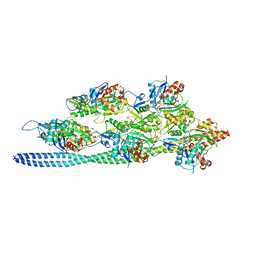 | | Structure of the F-actin-tropomyosin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | von der Ecken, J, Mueller, M, Lehman, W, Manstein, J.M, Penczek, A.P, Raunser, S. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the F-actin--tropomyosin complex.
Nature, 519, 2015
|
|
6FM7
 
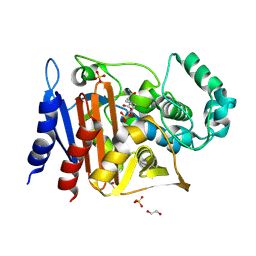 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Di Pisa, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
3PXU
 
 | |
2VUG
 
 | | The structure of an archaeal homodimeric RNA ligase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PAB1020, ... | | Authors: | Brooks, M.A, Meslet-Cladiere, L, Graille, M, Kuhn, J, Blondeau, K, Myllykallio, H, van Tilbeurgh, H. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of an archaeal homodimeric ligase which has RNA circularization activity.
Protein Sci., 17, 2008
|
|
4JAJ
 
 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6C41
 
 | |
6FM6
 
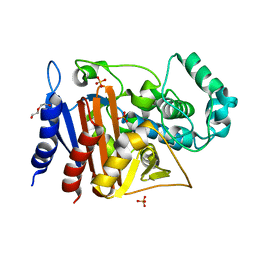 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes | | Descriptor: | Beta-lactamase, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Pozzi, C, De Luca, F, Di Pisa, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
6CEG
 
 | |
6FTX
 
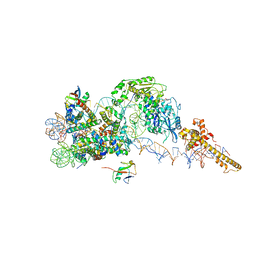 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
6BXD
 
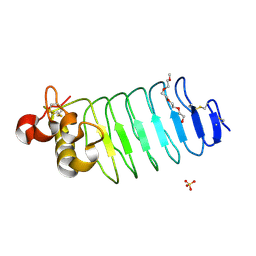 | | Crystal structure of Variable Lymphocyte Receptor 2 (VLR2) | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, SULFATE ION, Variable Lymphocyte Receptor 2 | | Authors: | Gunn, R.J, Wilson, I.A, Cooper, M.D, Herrin, B.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.103 Å) | | Cite: | VLR Recognition of TLR5 Expands the Molecular Characterization of Protein Antigen Binding by Non-Ig-based Antibodies.
J. Mol. Biol., 430, 2018
|
|
3P83
 
 | | Structure of the PCNA:RNase HII complex from Archaeoglobus fulgidus. | | Descriptor: | DNA polymerase sliding clamp, Ribonuclease HII | | Authors: | Bubeck, D, Reijns, M.A, Graham, S.C, Astell, K.R, Jones, E.Y, Jackson, A.P. | | Deposit date: | 2010-10-13 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | PCNA directs type 2 RNase H activity on DNA replication and repair substrates.
Nucleic Acids Res., 39, 2011
|
|
3HPA
 
 | | Crystal structure of an amidohydrolase gi:44264246 from an evironmental sample of sargasso sea | | Descriptor: | AMIDOHYDROLASE, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The hunt for 8-oxoguanine deaminase.
J.Am.Chem.Soc., 132, 2010
|
|
2WBE
 
 | | Kinesin-5-Tubulin Complex with AMPPNP | | Descriptor: | BIPOLAR KINESIN KRP-130, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bodey, A.J, Kikkawa, M, Moores, C.A. | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | 9-Angstrom Structure of a Microtubule-Bound Mitotic Motor.
J.Mol.Biol., 388, 2009
|
|
